How to Use the Bispecific Module in PKSim: Step-by-Step Demo & Instructions
Learn how to use the bispecific module in PKSim, including renaming molecules, handling reactions, and building simulations for bispecific drugs targeting PD-1 and TNF. Follow this step-by-step guide for efficient model extension and quality checks.
In this guide, we'll learn how to use the bispecific extension module in a PKSim model to add features such as trimer formation, target turnover, internalization, recycling, and degradation for each target and complex.
We'll focus on the process of renaming and removing molecules and reactions to ensure the module works correctly with your base model. This will help you prepare your model for simulation and quality checks.
Let's get started
Okay. Here is a quick demo and instructions on how to use the bispecific module we created. The expected starting point for this extension module is a bispecific PKsim model, which already includes most of the relevant molecules.
We have a bispecific drug that binds to two different targets. In this case, we made it PD-1 and TNF. We have the complex formation in the base model, but that's all.
We need to include trimer formation, as well as the turnover of all targets: internalization, recycling, and degradation for each target and each complex.
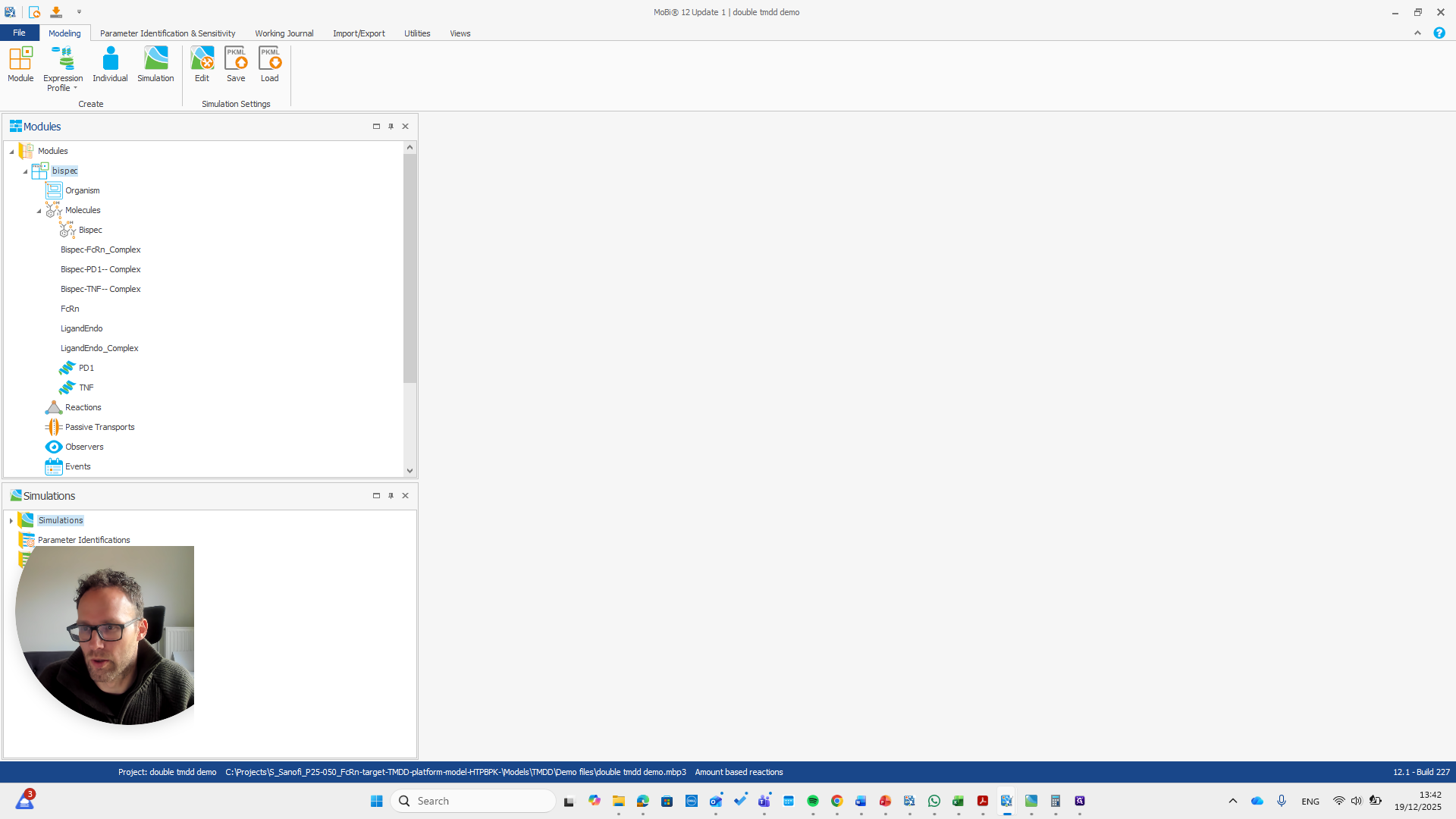
That is what we do with the extension module here.

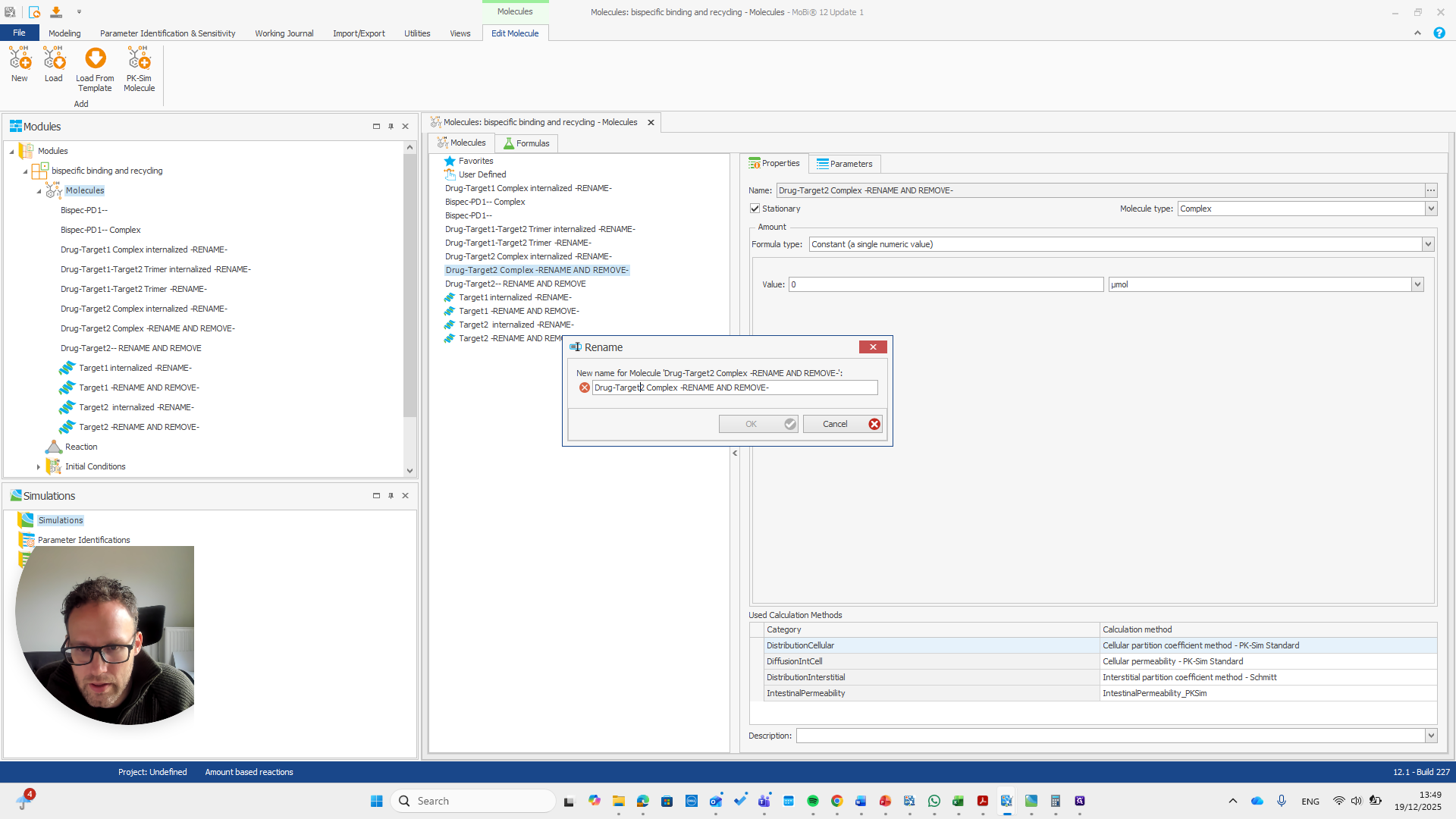
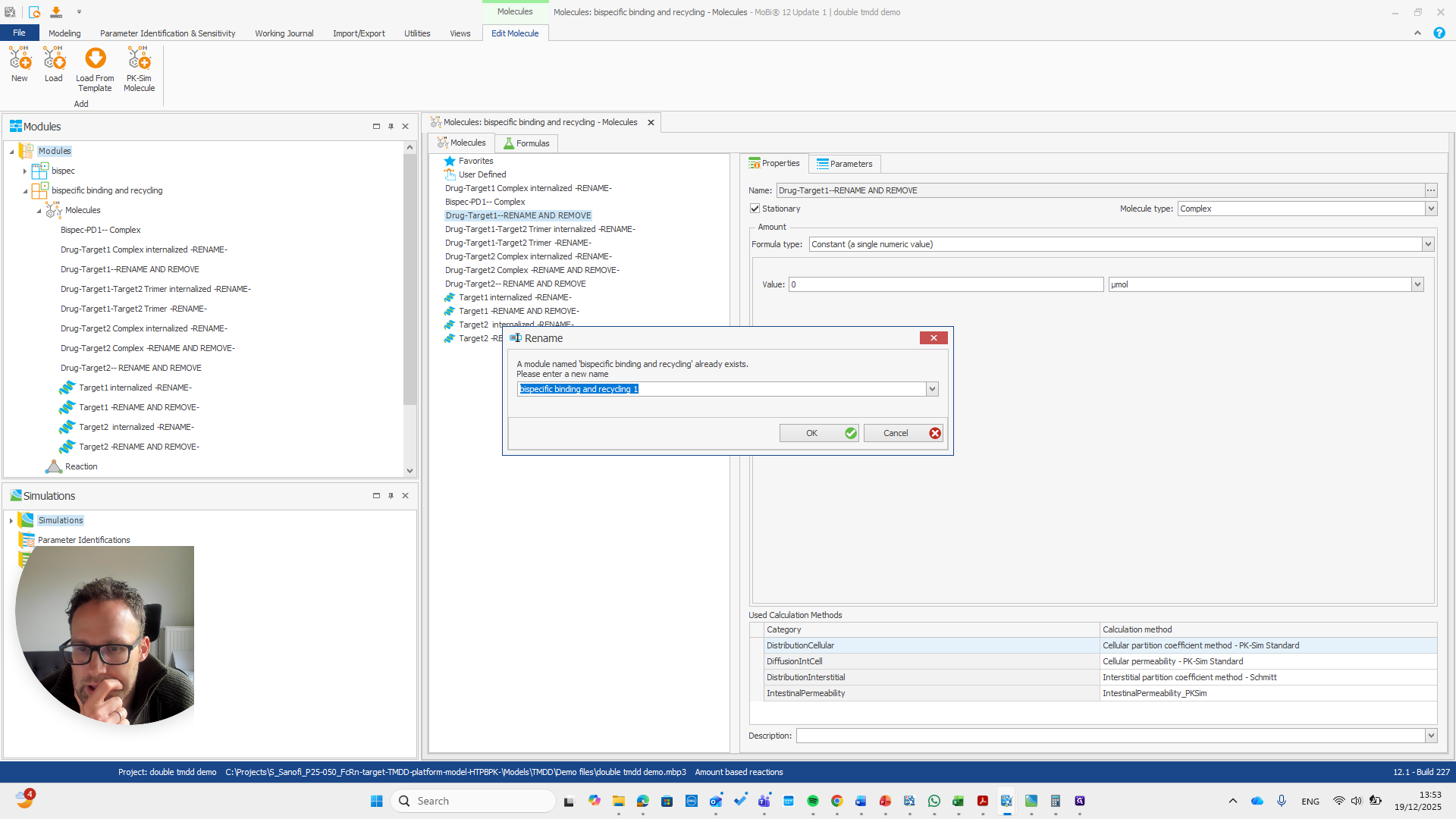
We have a molecule building block that needs to be renamed.

It's important to note that only the "rename and remove" actions need to be completed. The others are mainly for your convenience, as well as for proper transparency and documentation. I will now demonstrate renaming this using only the rename and remove options. This approach makes the process shorter.
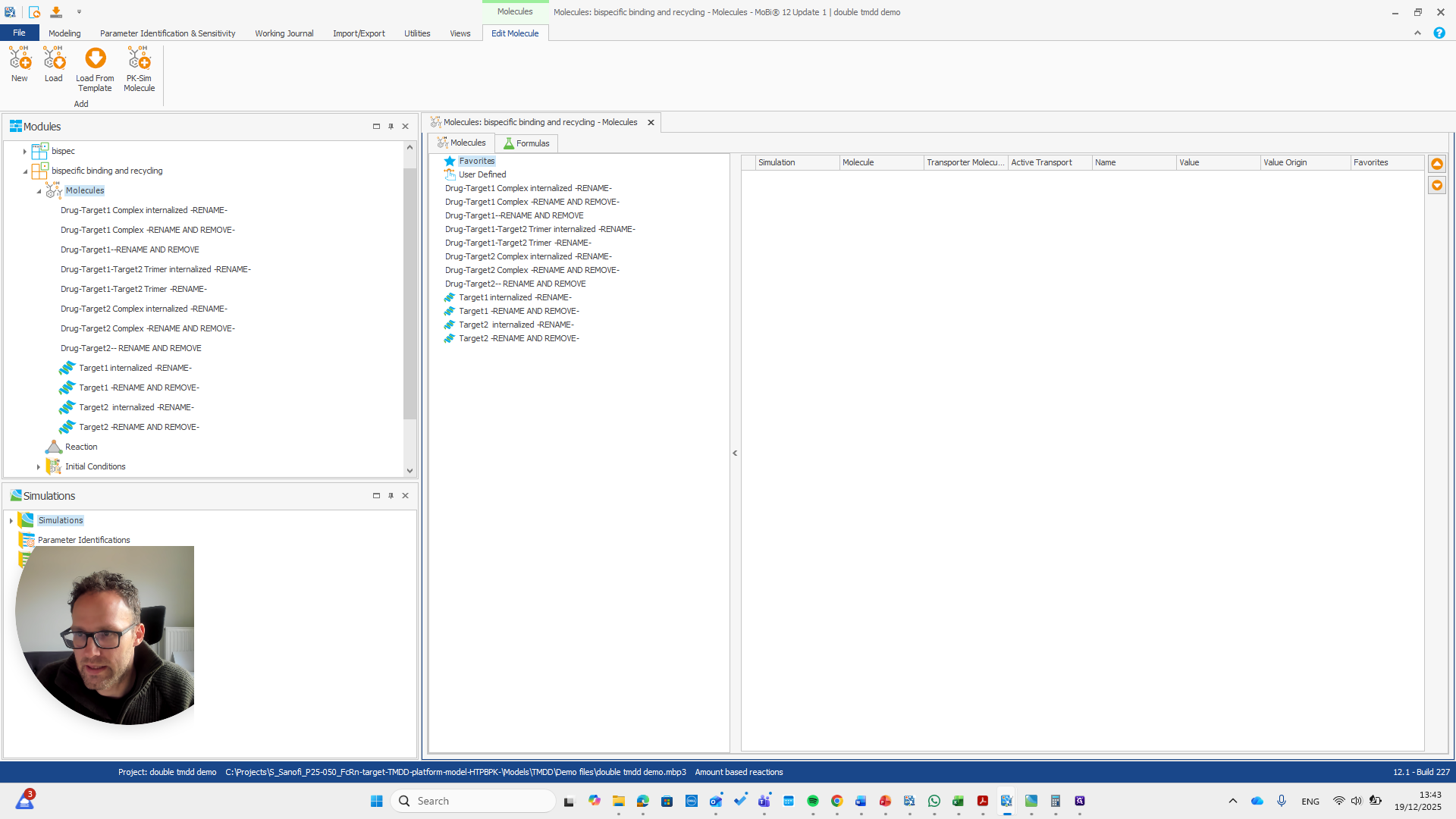
We need to pay attention because all these molecules are also in the base module. One molecule here is named after a reaction. This is simply a way to rename a reaction and its pathway for a parameter.
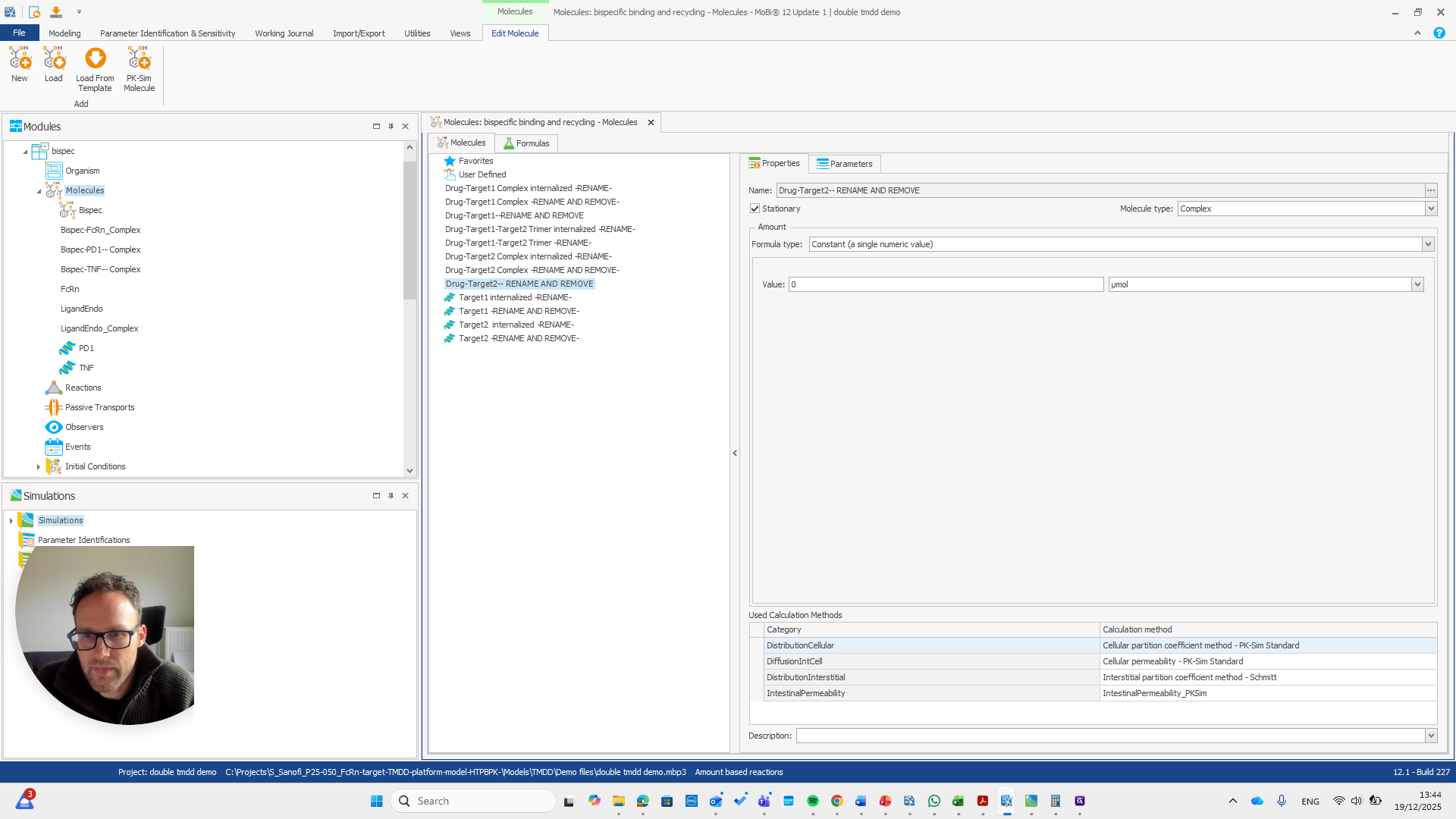
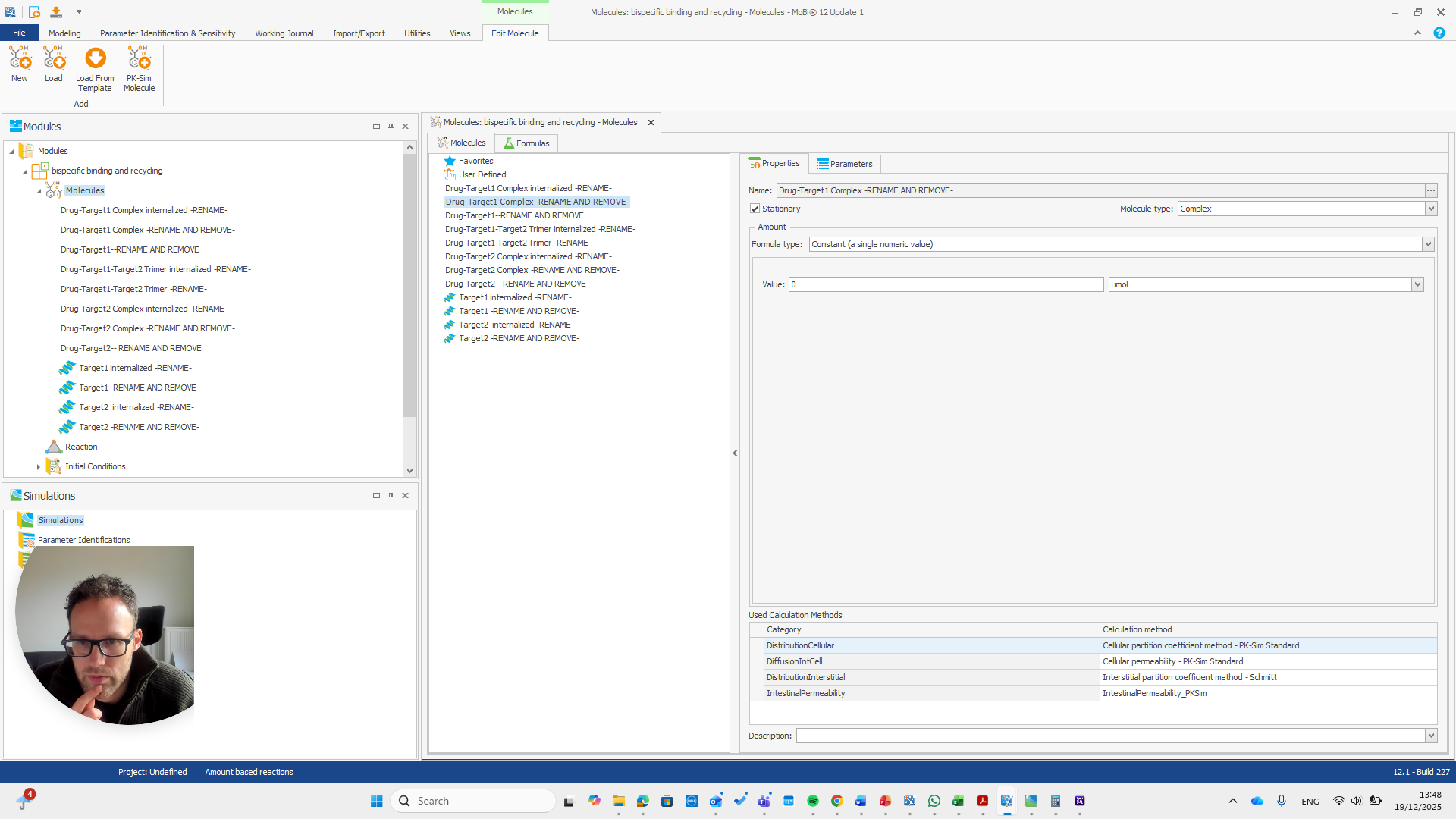
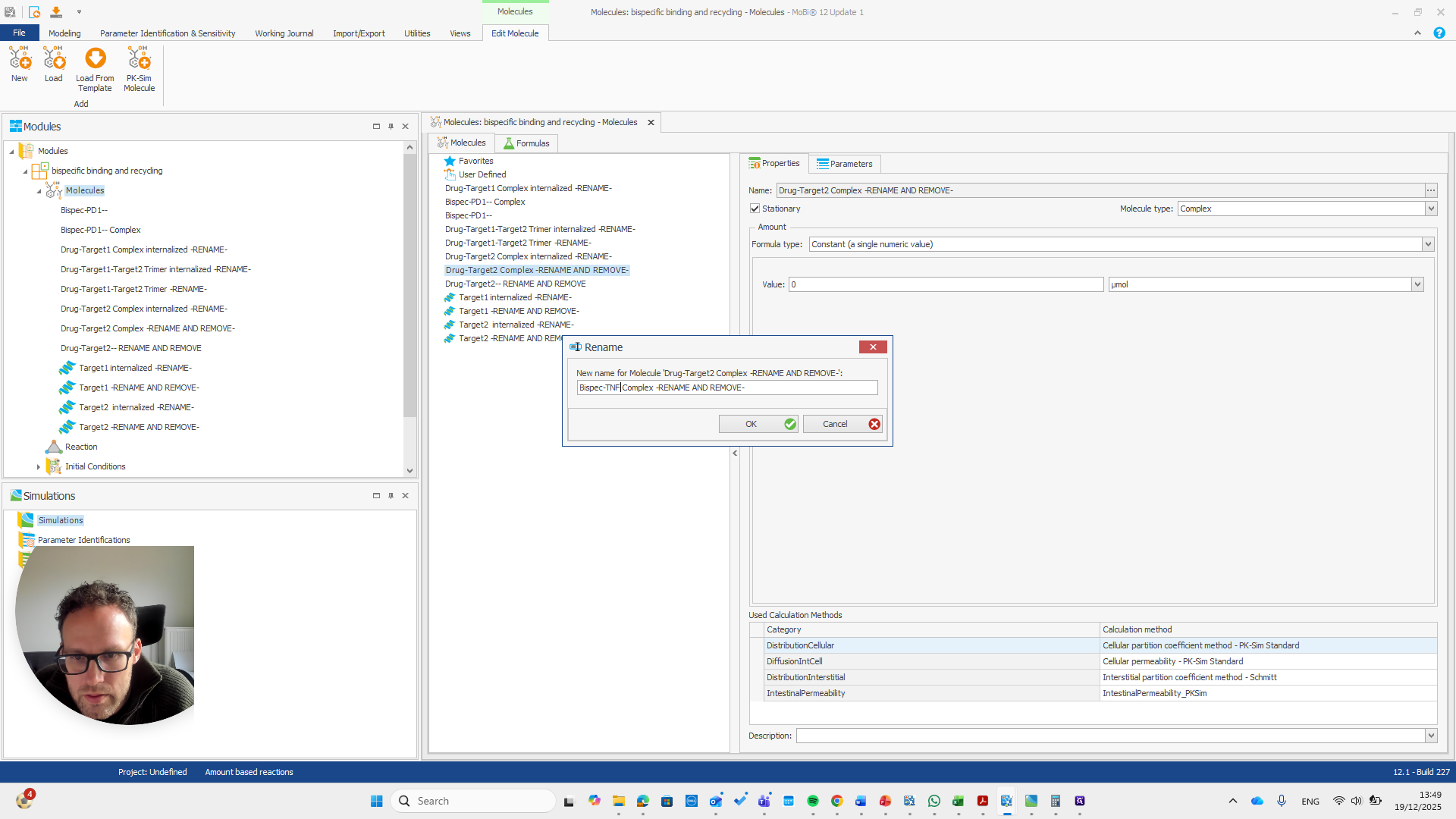
Drug target one complex needs to be renamed to bispec PD1 complex.
We can do that here.
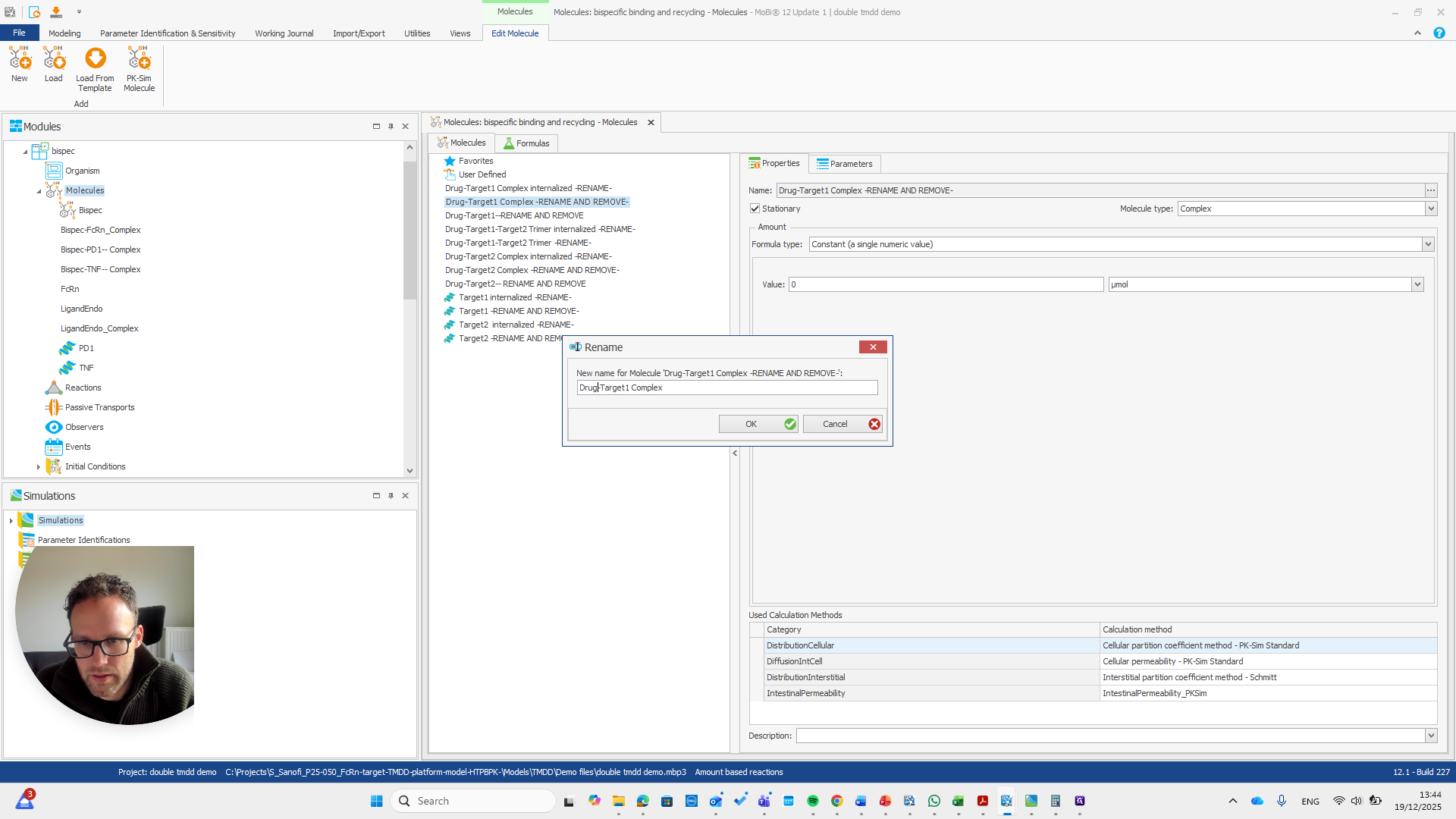
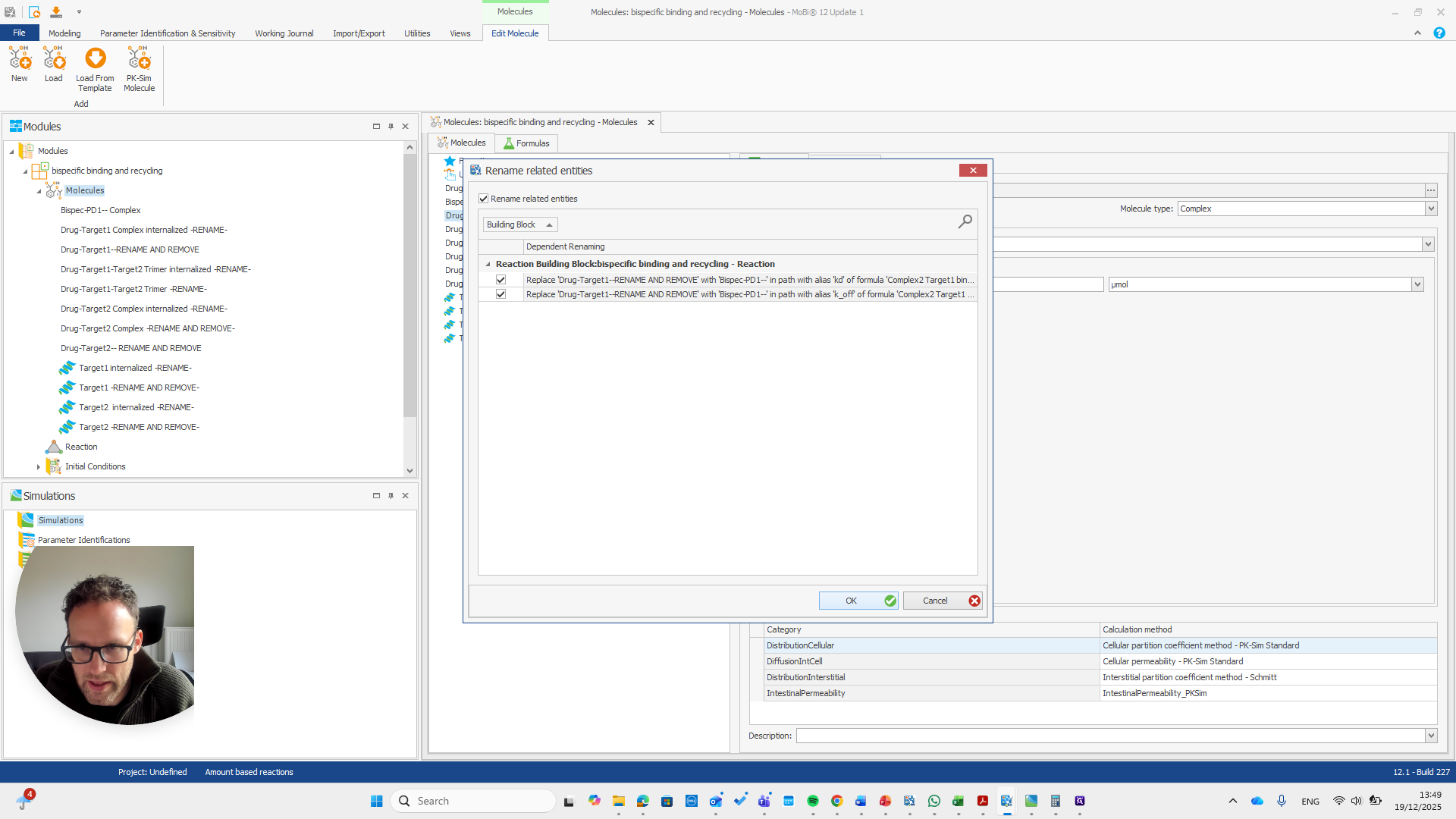
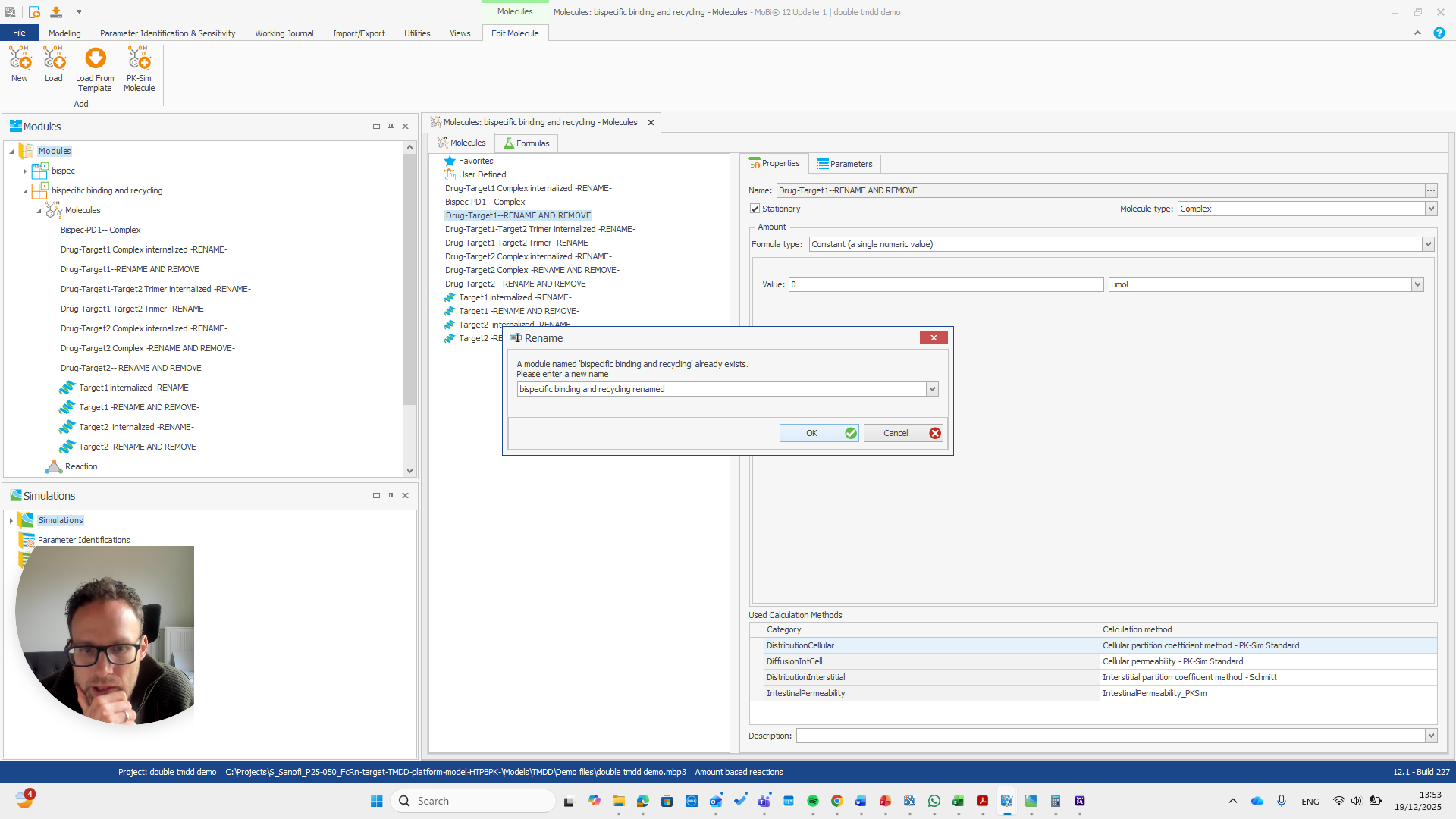
We automatically receive a request if everything needs to be renamed, which is the case here.
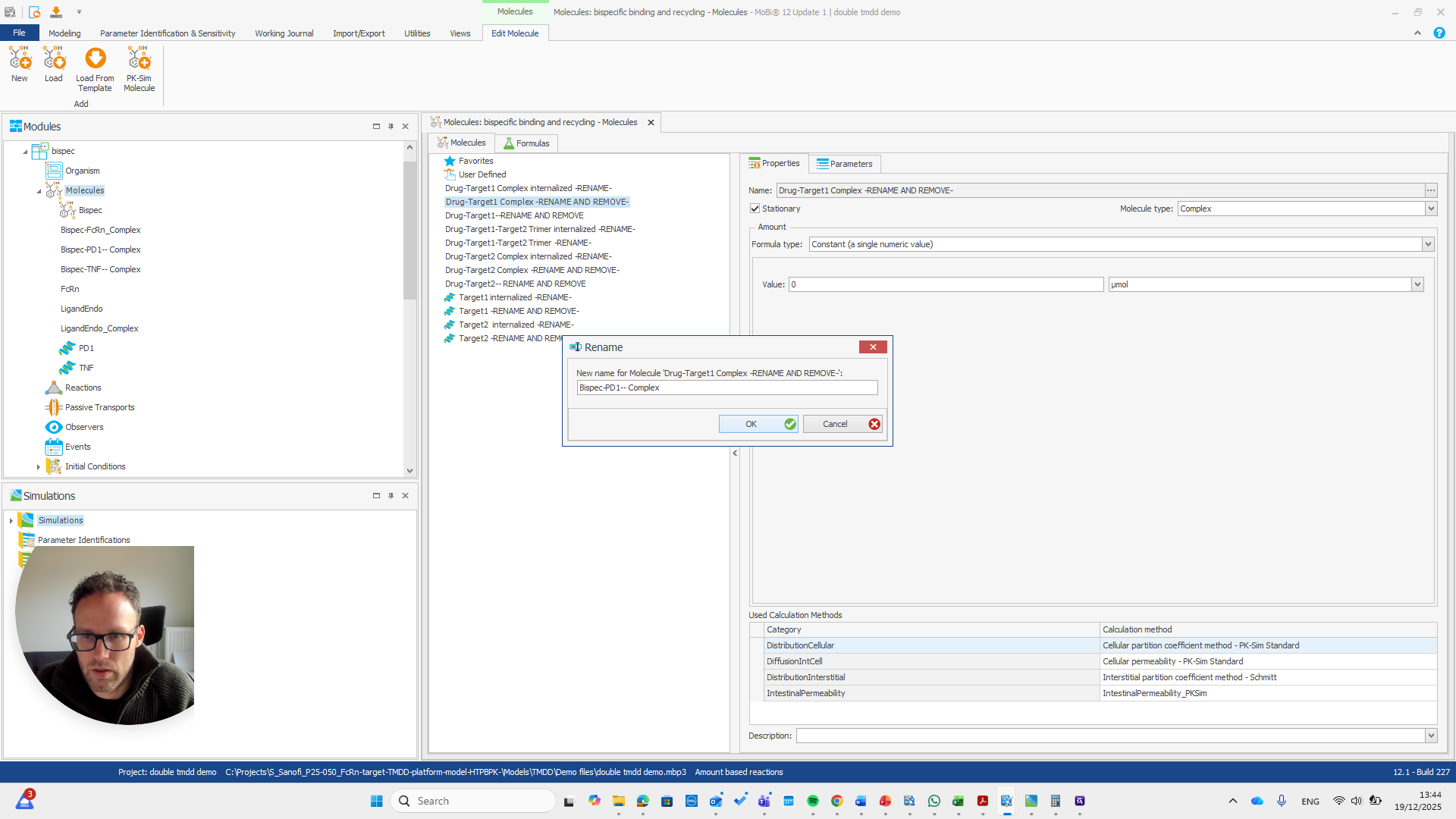
I will double-check that the name and spelling match exactly with the molecule's name. Yeah.
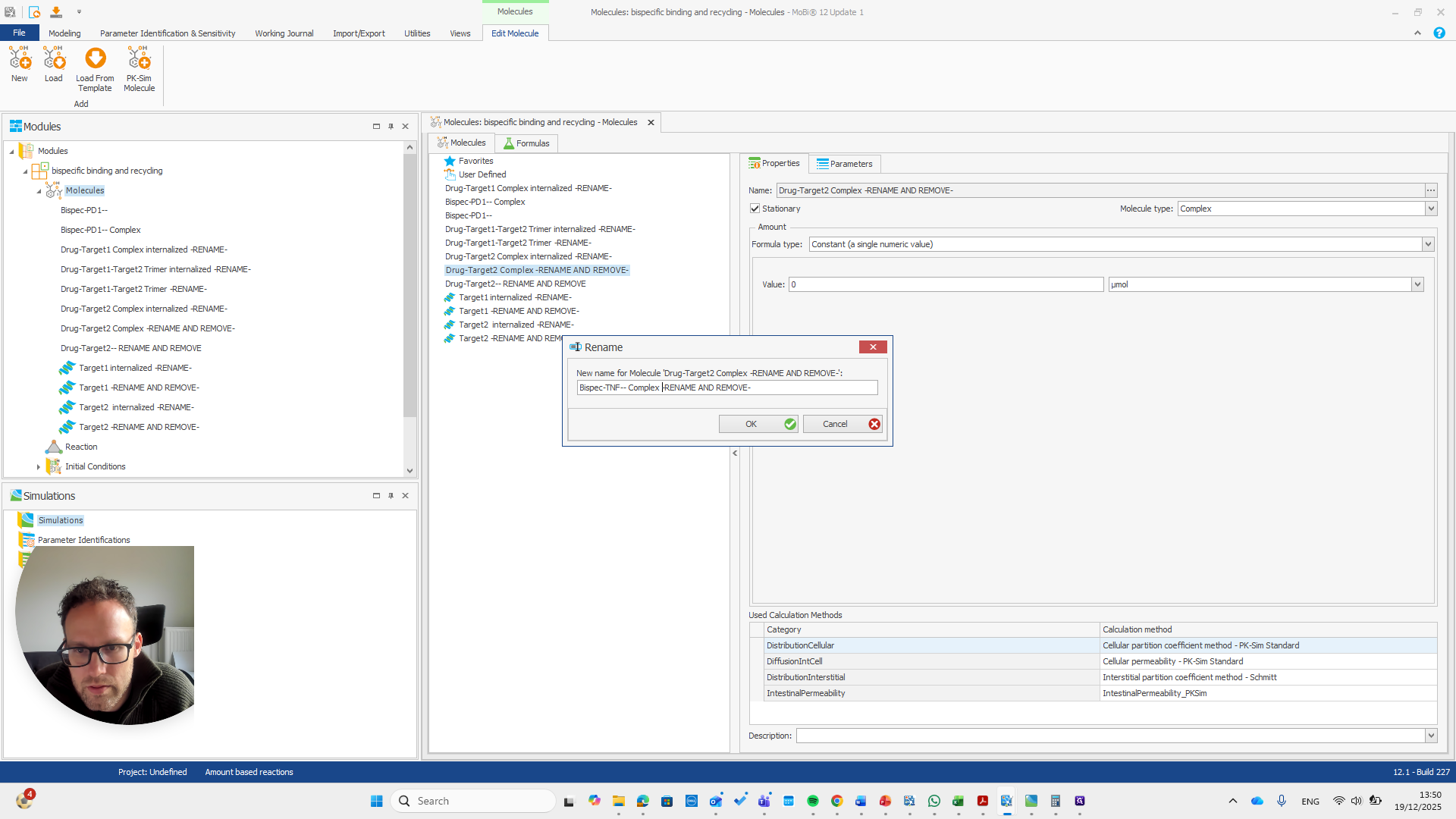
This is the other reaction. There are actually two reactions here.
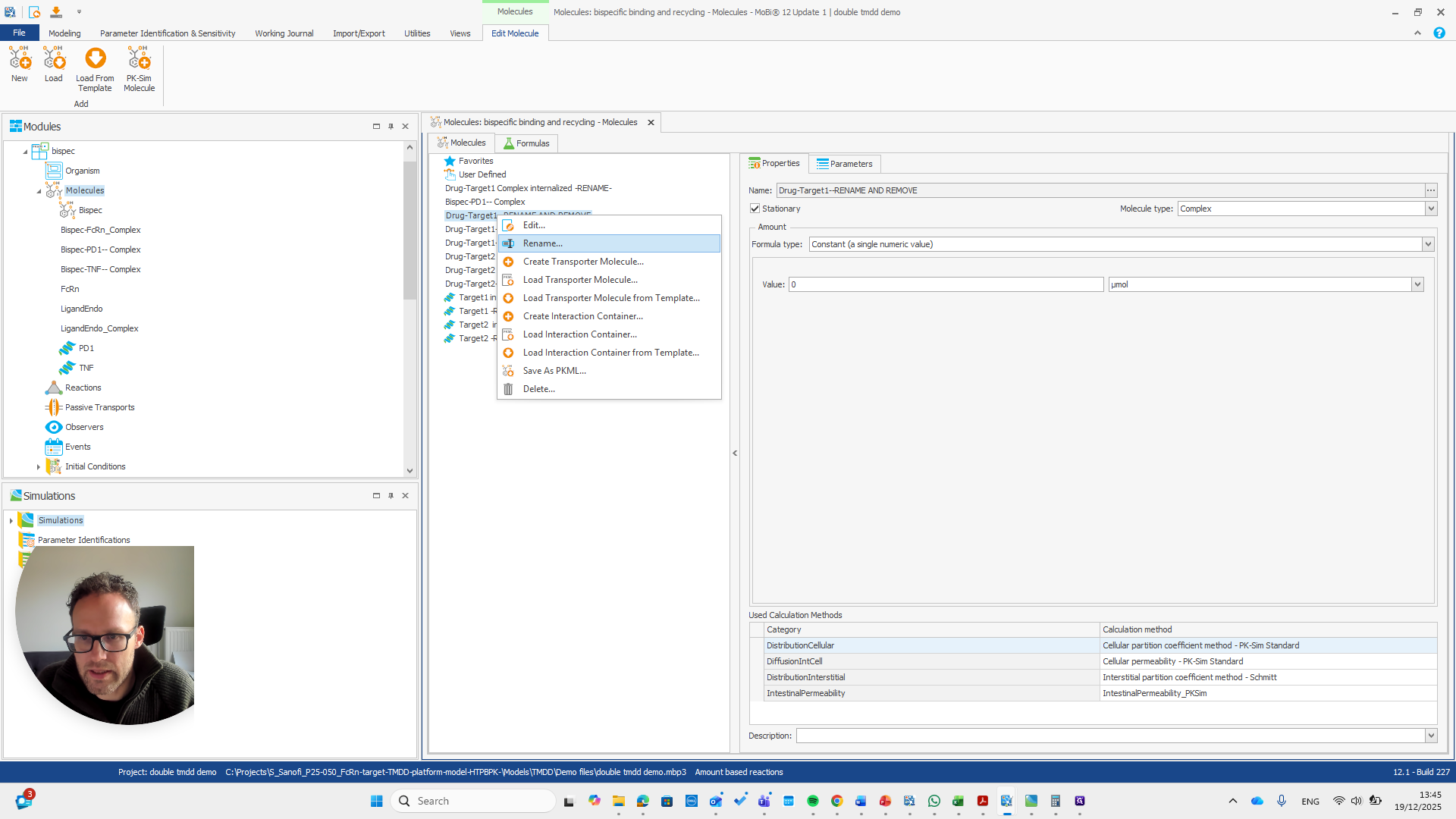
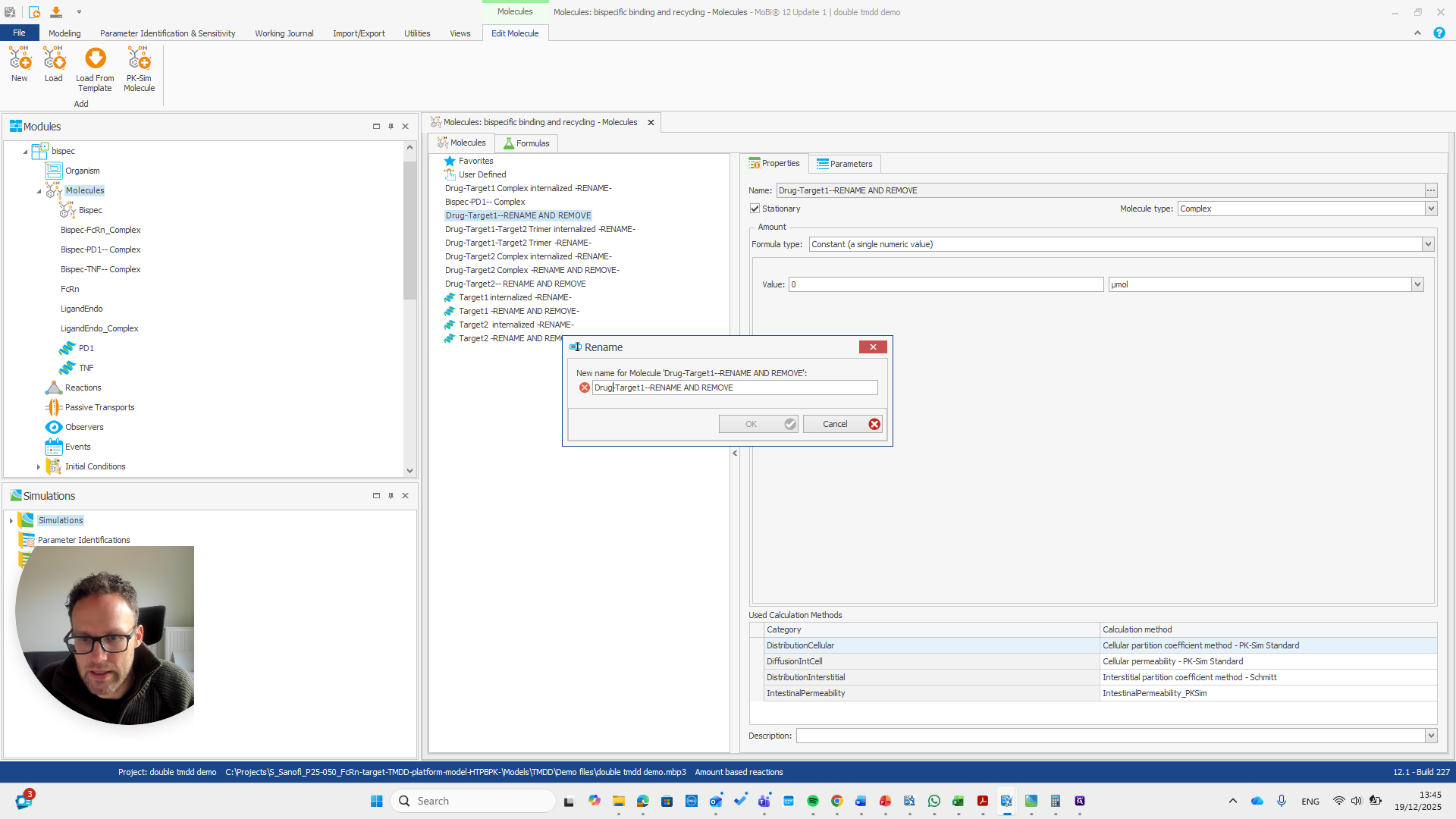
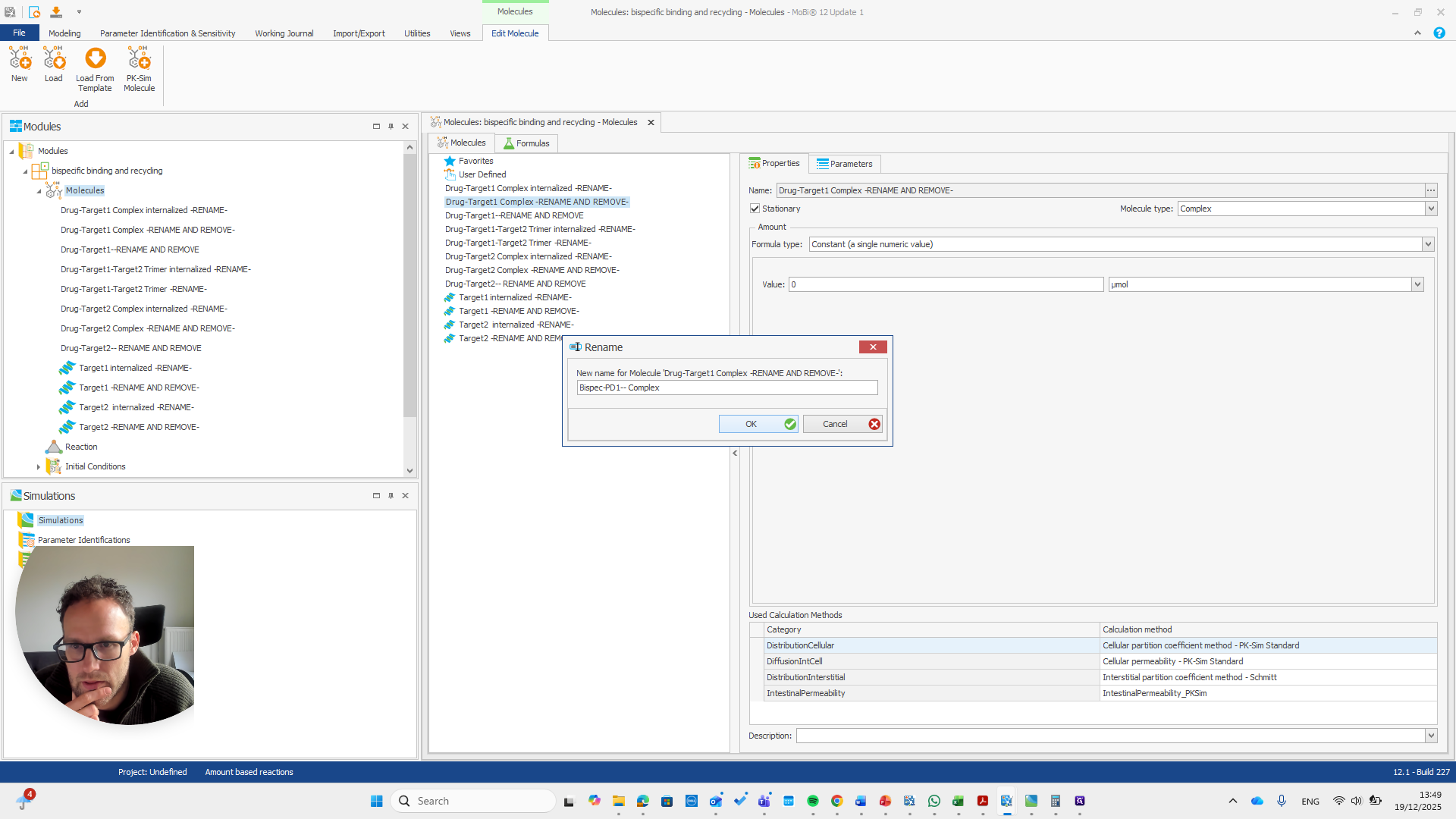
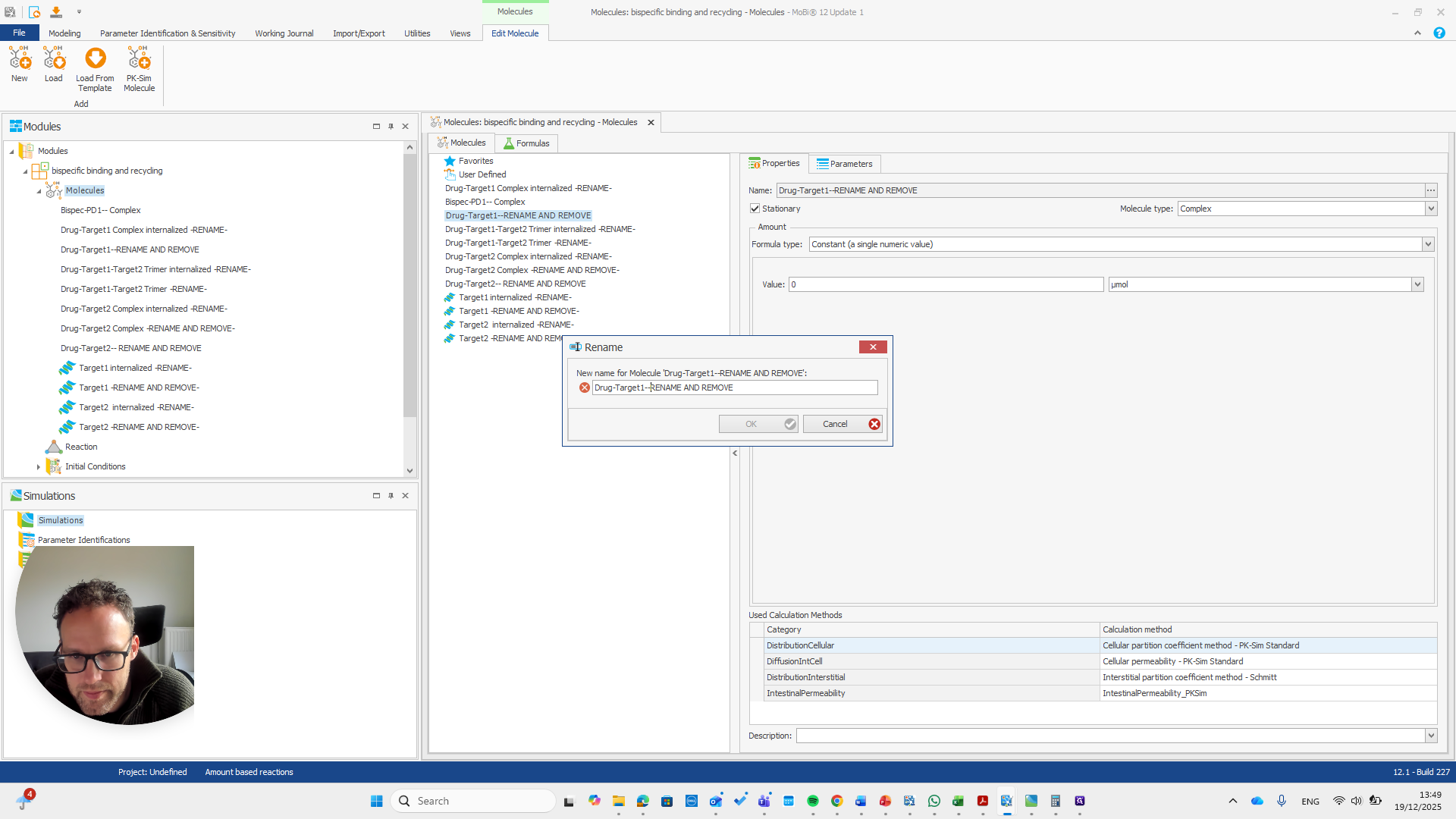
So should be "bispec PD1", followed by a double dash, and nothing else.
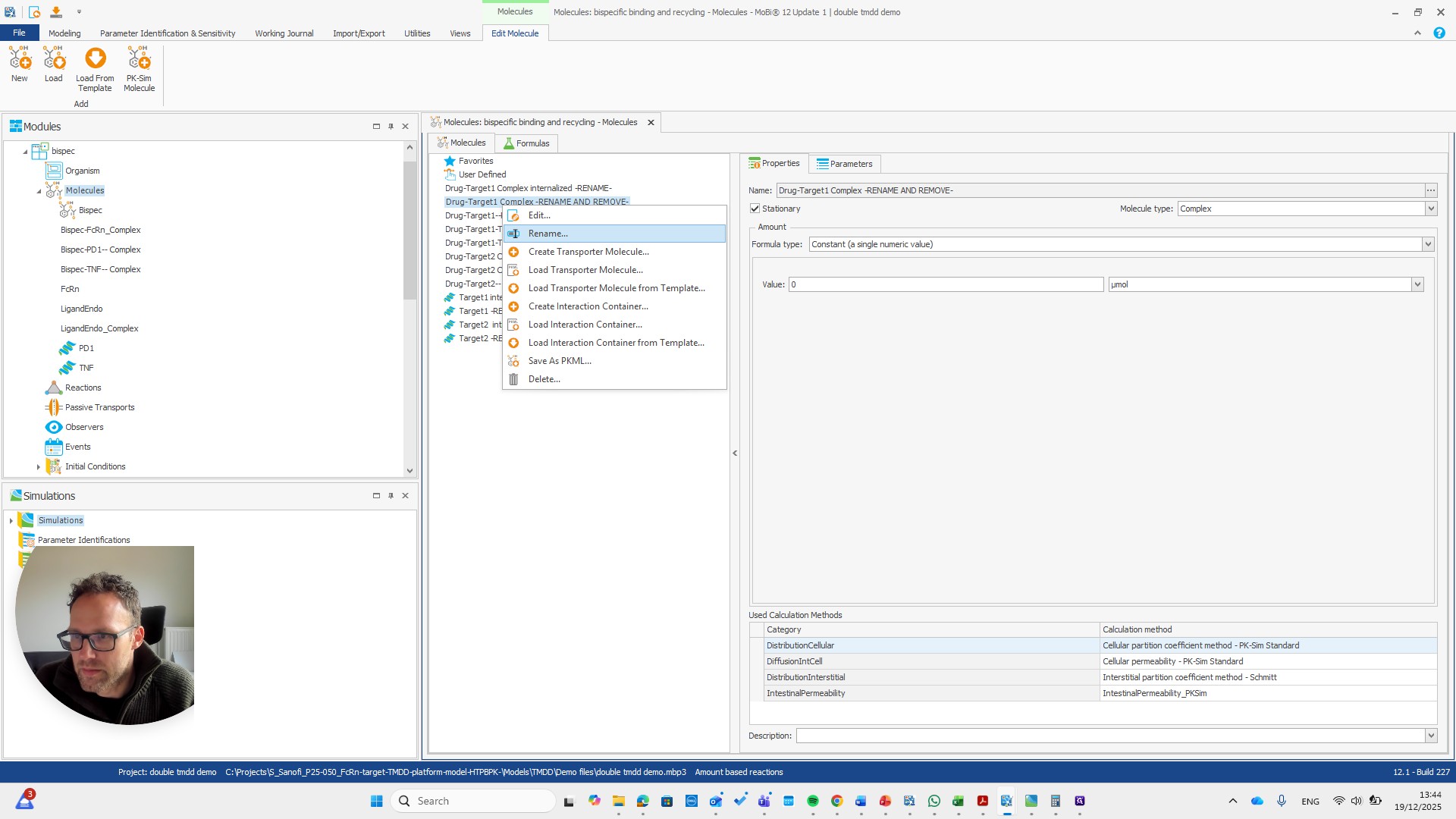
This issue occurs regularly. The problem is that we cannot rename it to... We cannot rename this reaction or molecule to the name of a reaction because MOBI recognizes that a reaction with that name already exists. It assumes this is not what we want and does not allow the change. We need to open a new MOBI file and rename it there. Okay.
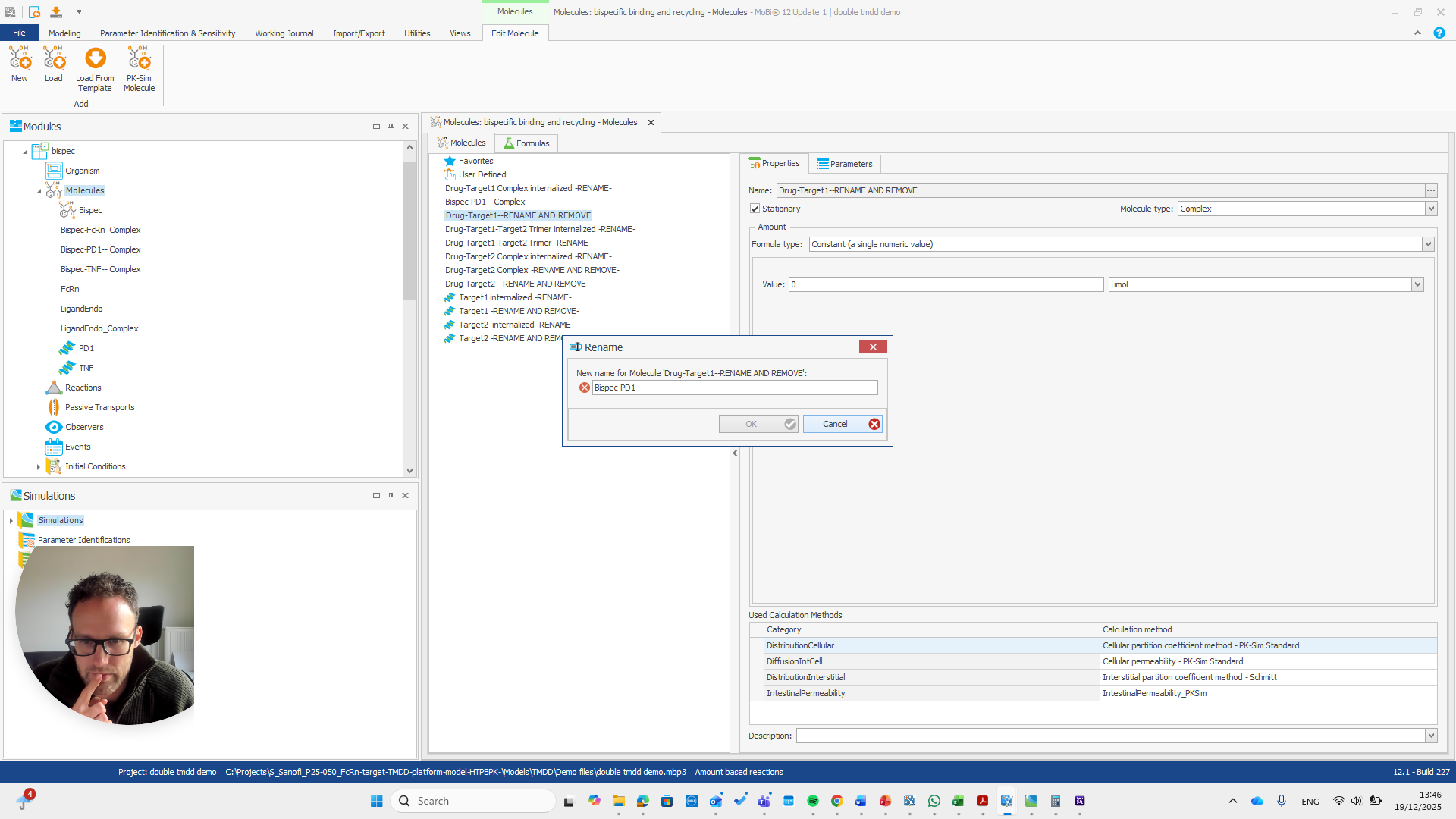
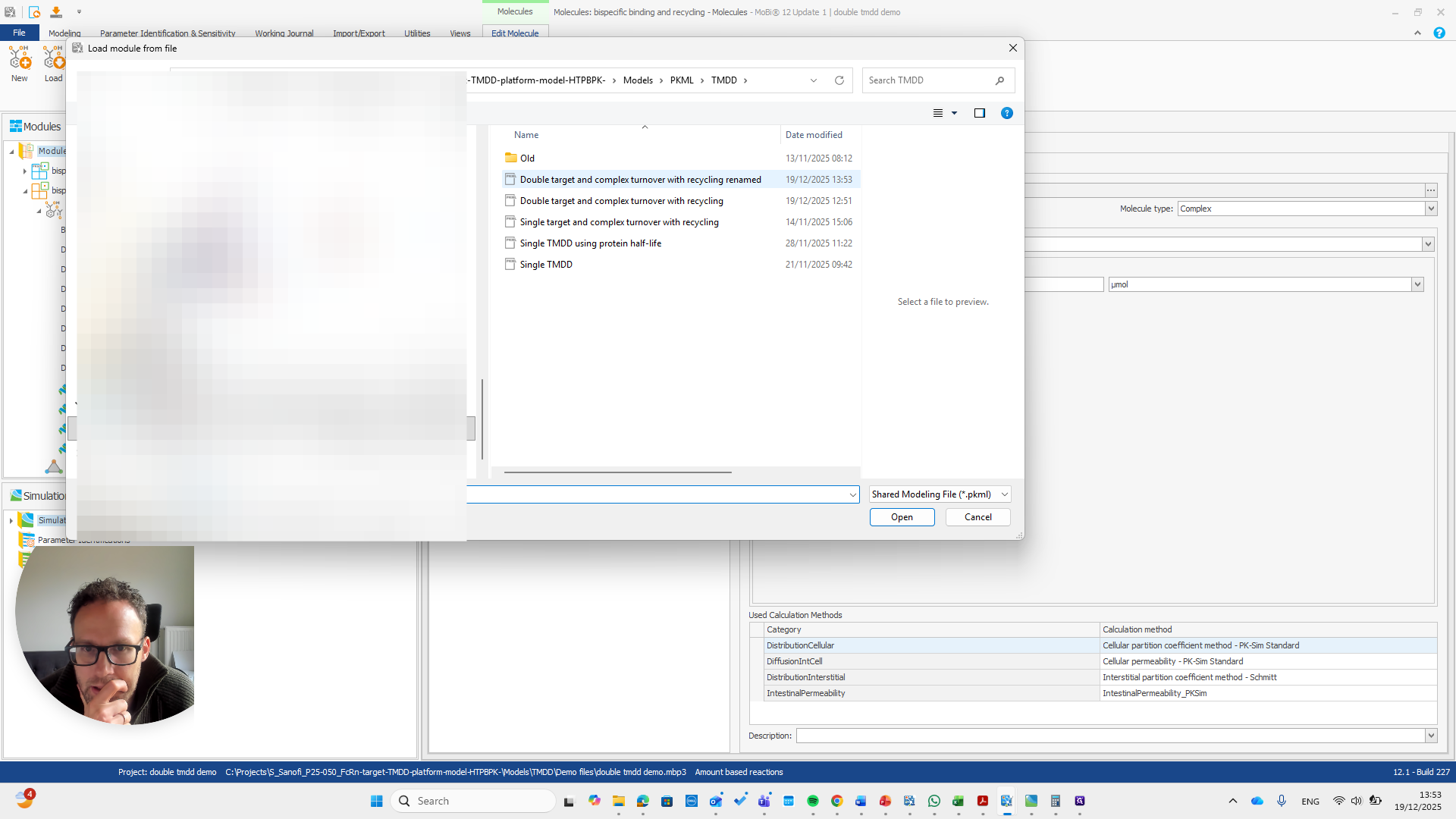
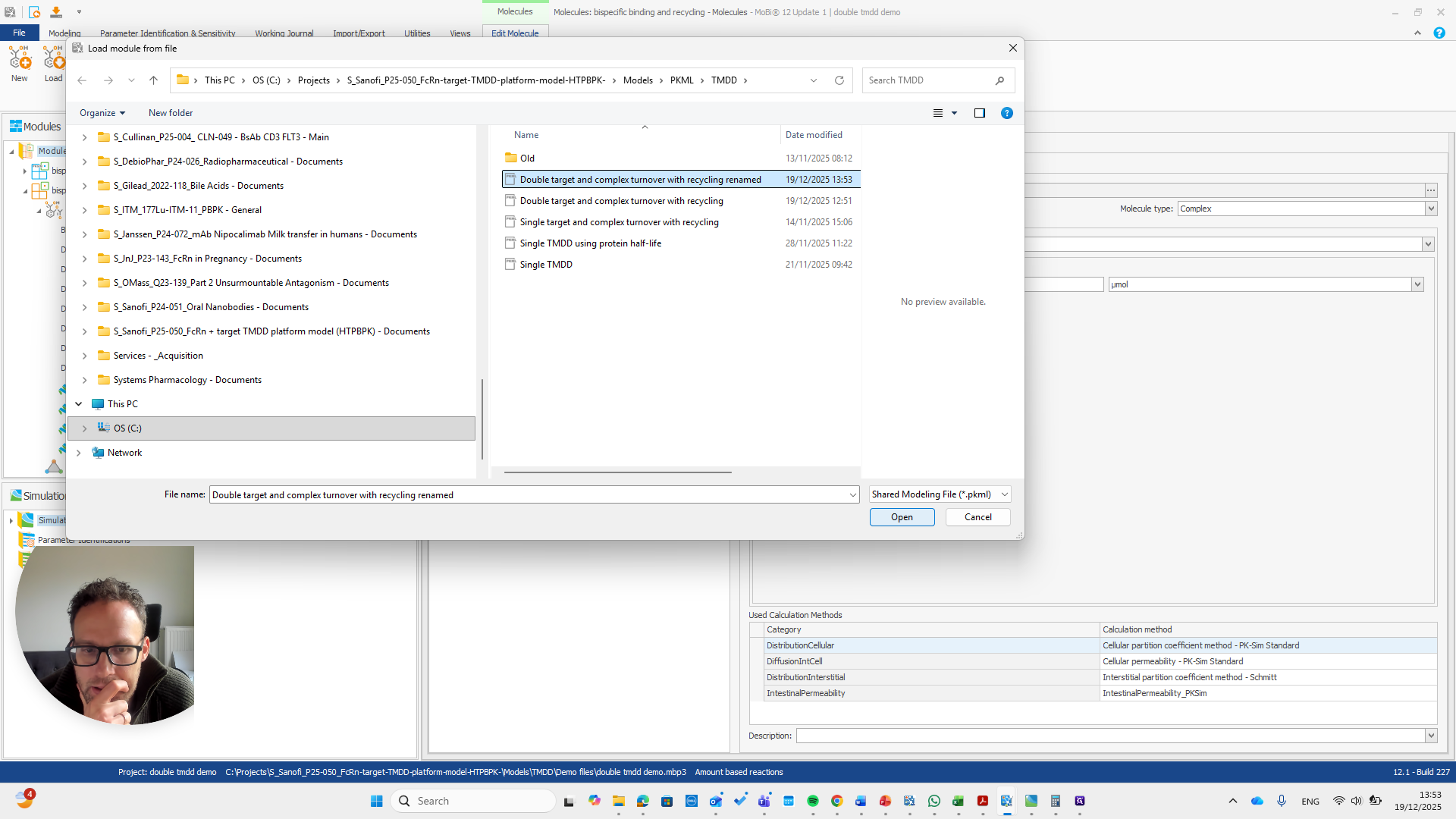
We open a new MOBI file and import the module.
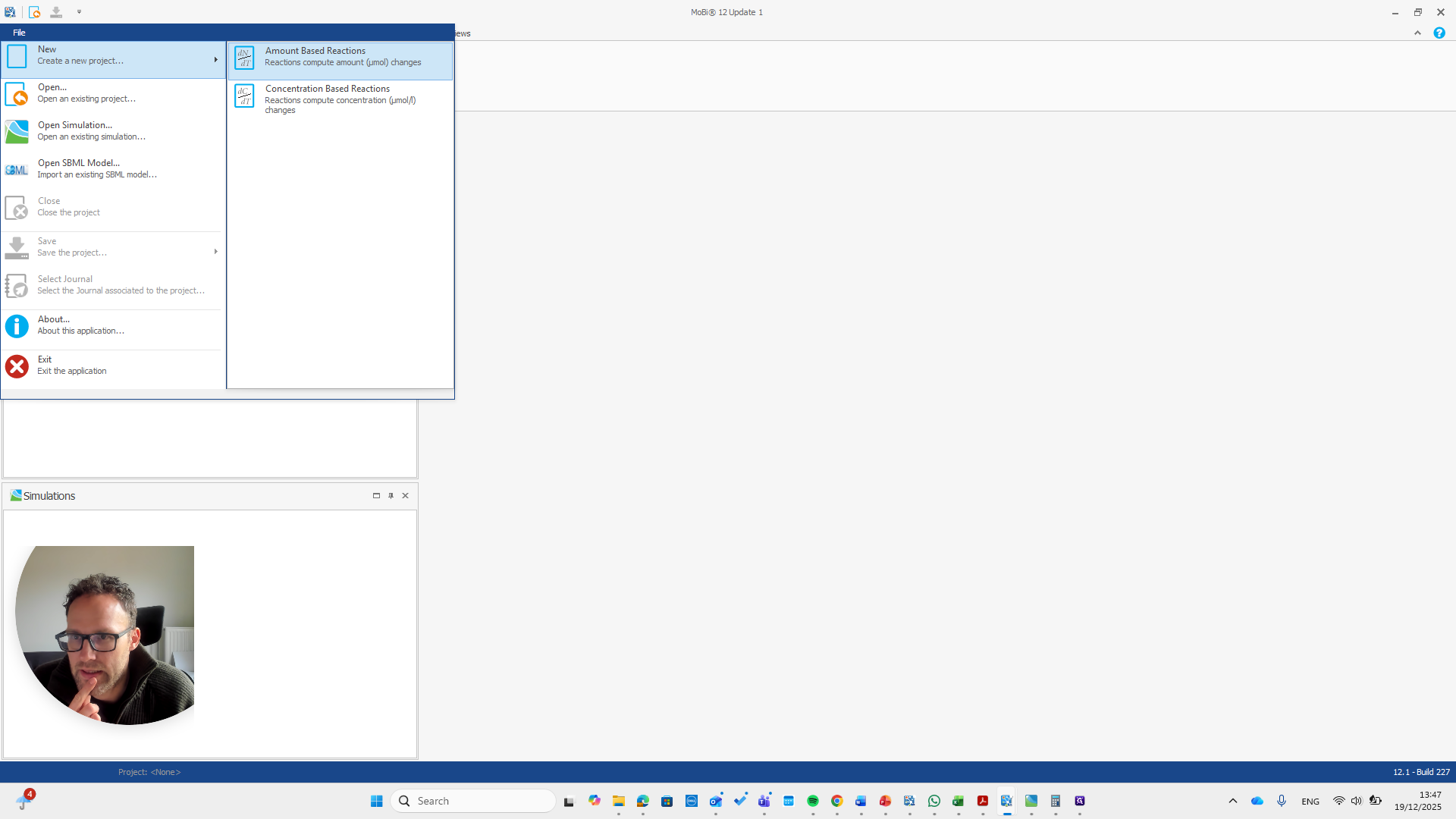
First, we need to state that this is an amount-based reactions model. Only then can we load the module.
The module is now loaded. We can repeat the same process.
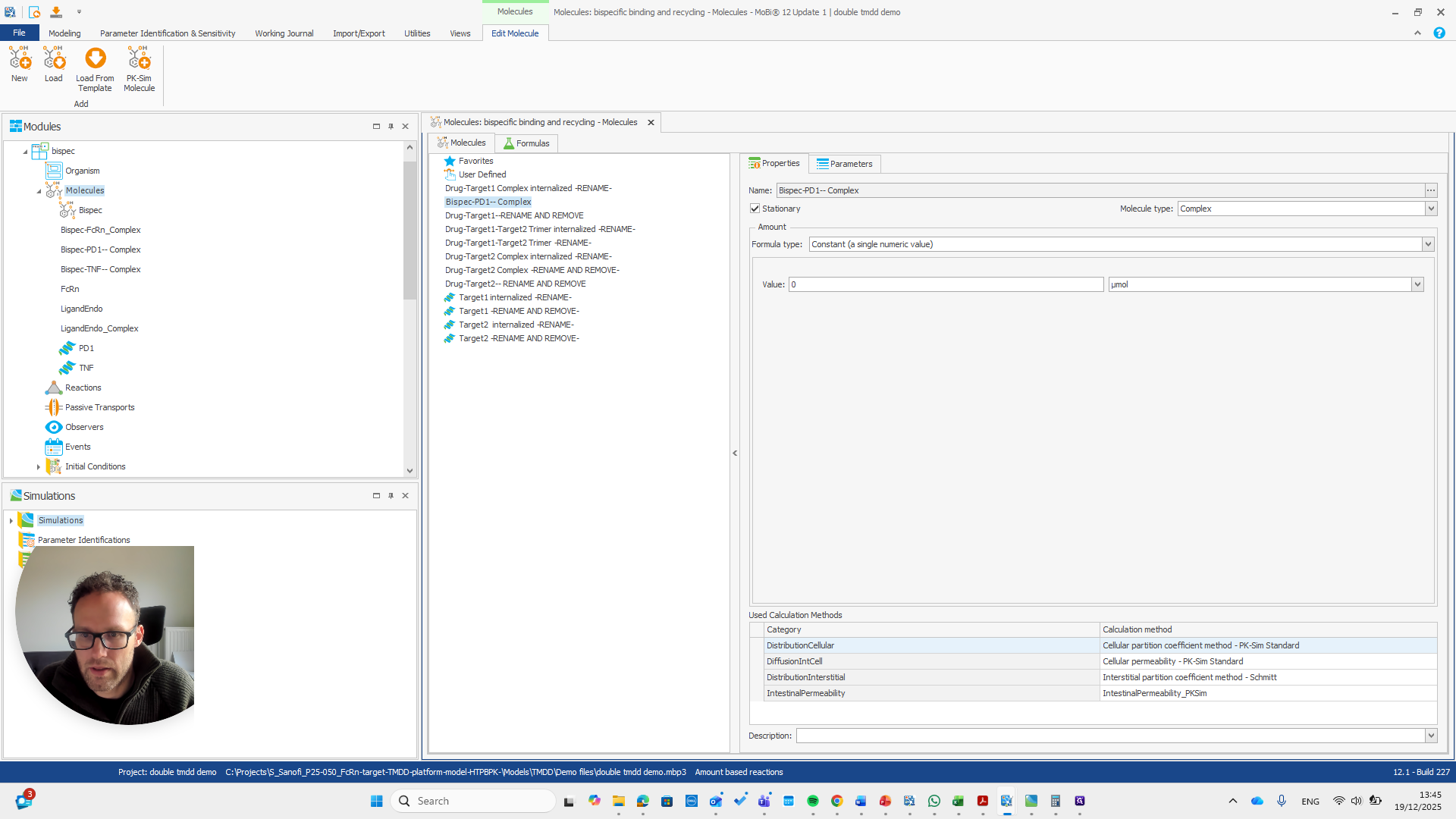
This is a bispecific PD1 complex.

You can check your other Moby file to confirm if this is what you need.
Okay.
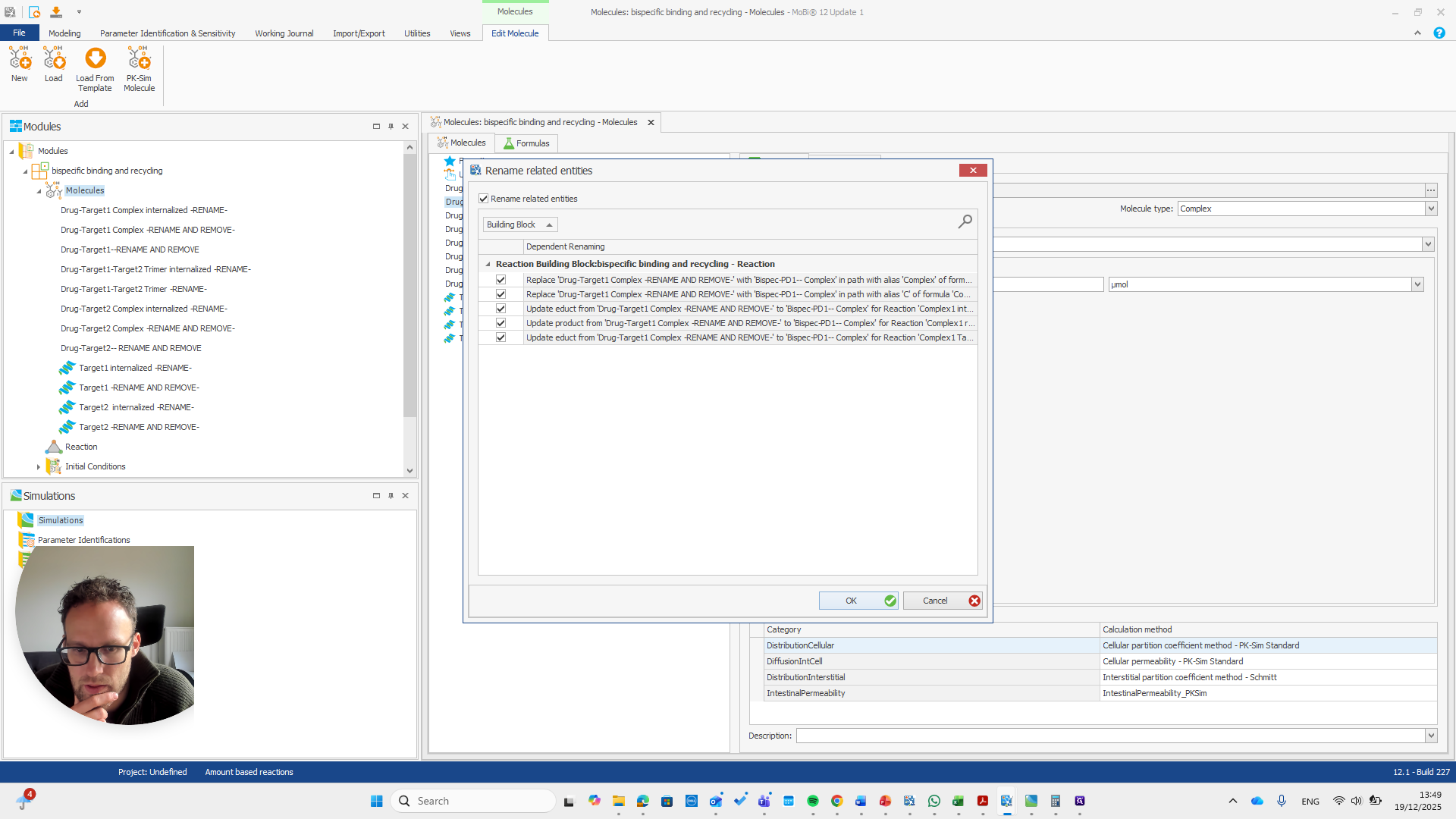
This, again, is for the reaction.

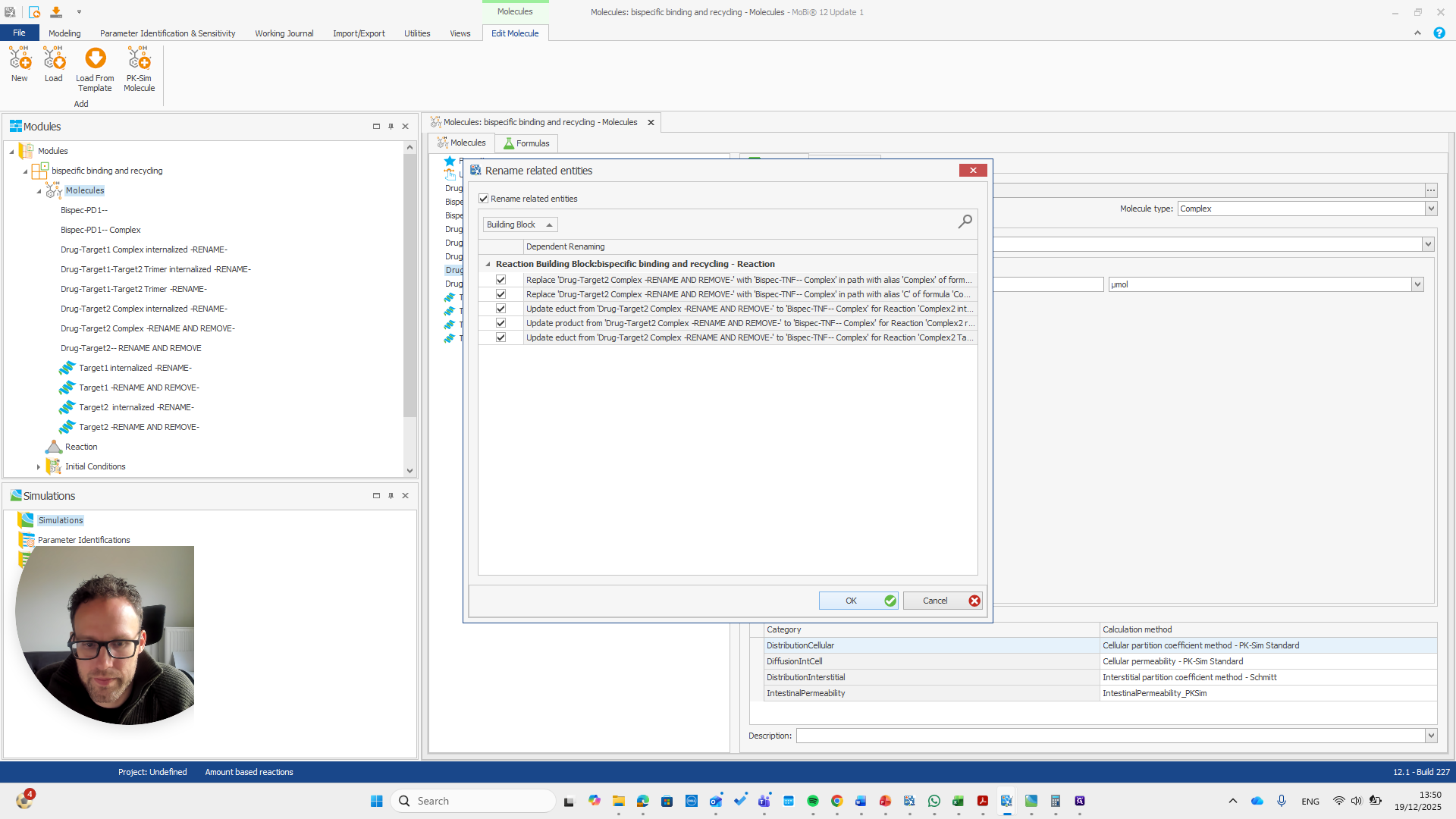
If you do it correctly, you will see that it also finds some paths that will be renamed with it.
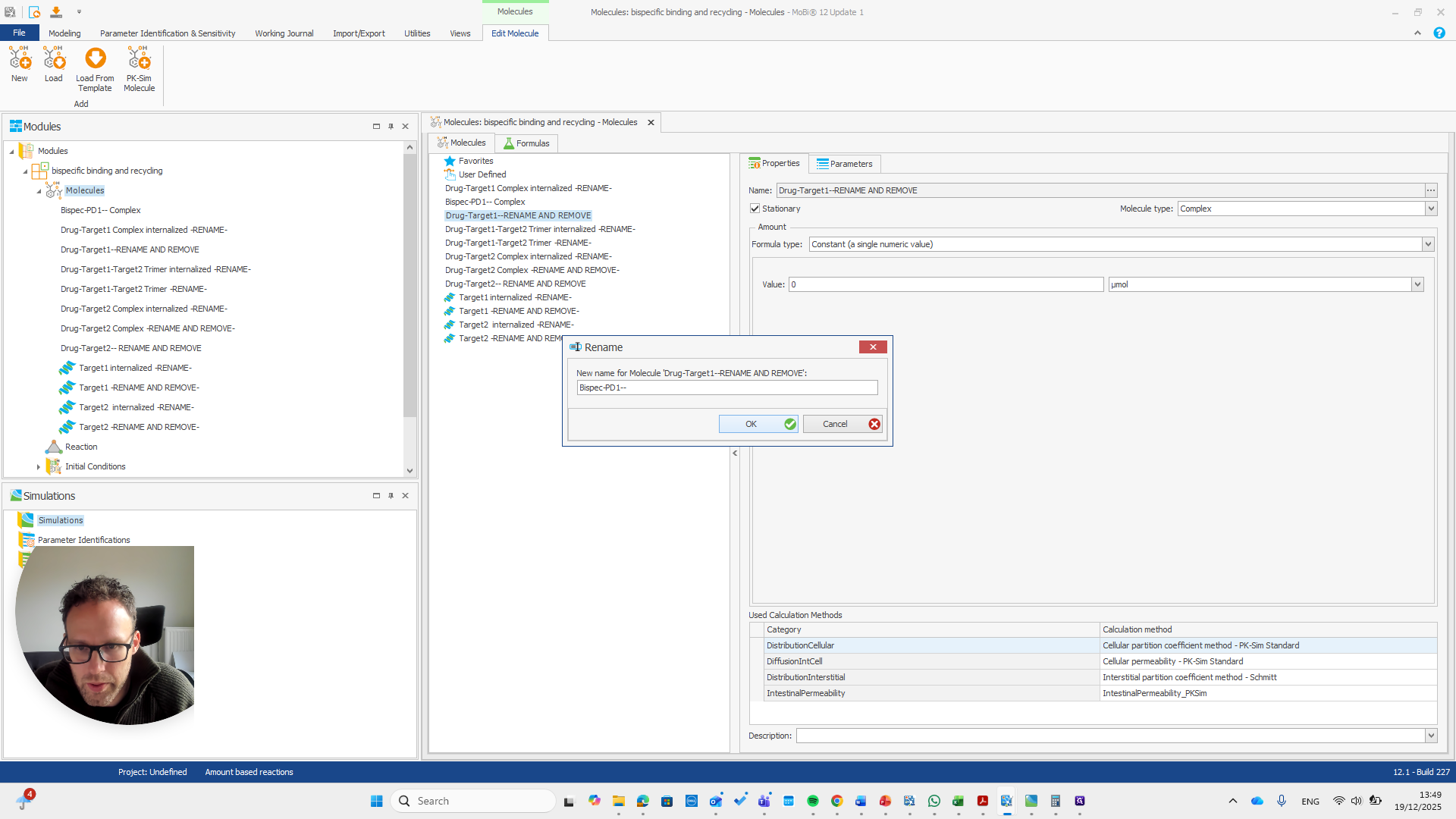
If you do not see this pop-up, you have not entered the name correctly.
Okay.
This is now the TNF target.
There we go.
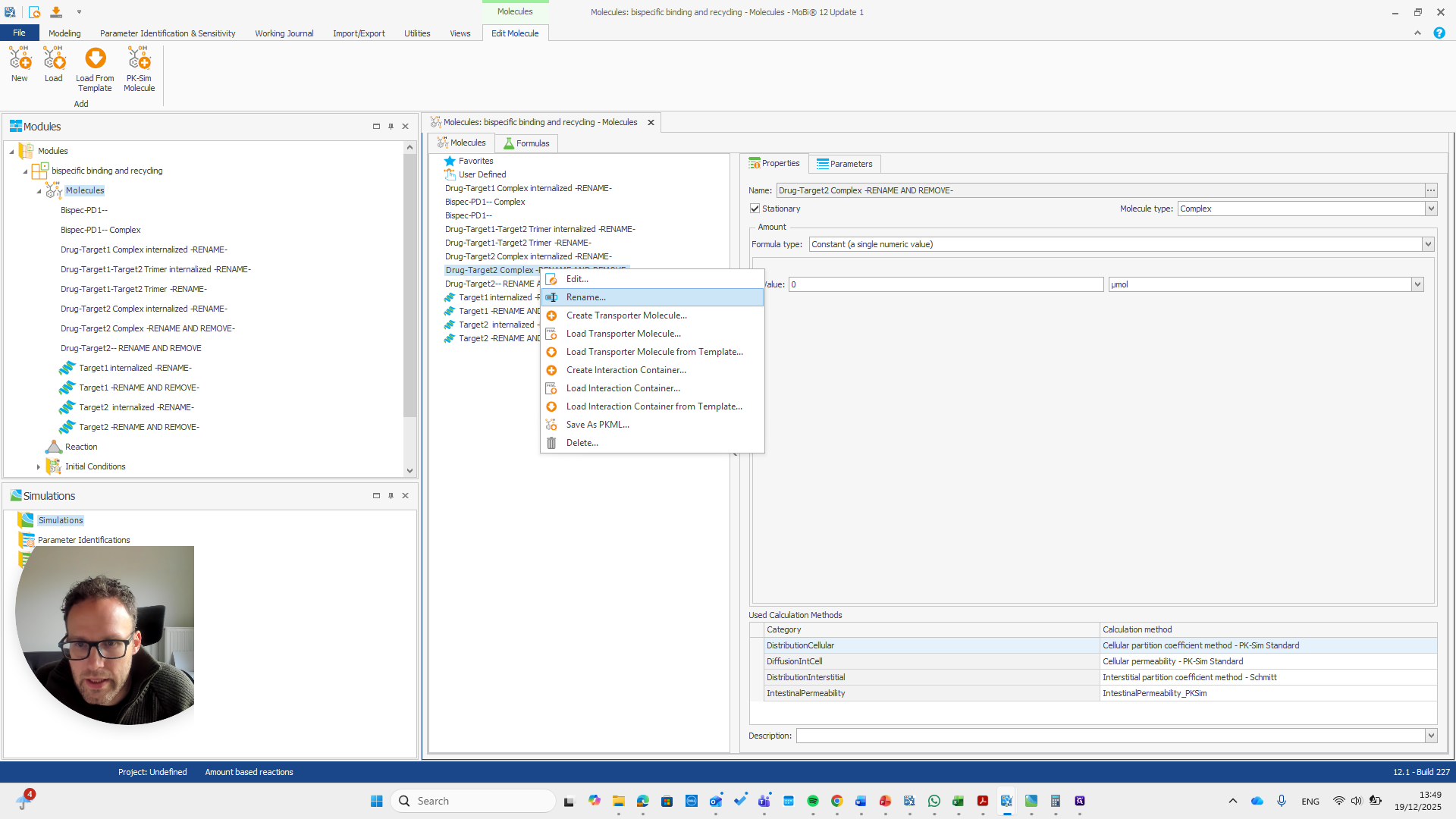
repeat this for all molecules with -RENAME AND REMOVE- and remove them all after renaming
Now, all the other molecules here did not need to be renamed or removed.
That is why they do not need to be renamed, since there is nothing they need to match. These are all new molecules, such as the trimer or any internalized target and drug target complexes. Okay. Now we have the module with the new name for our base module. We can save this as a PKML file.
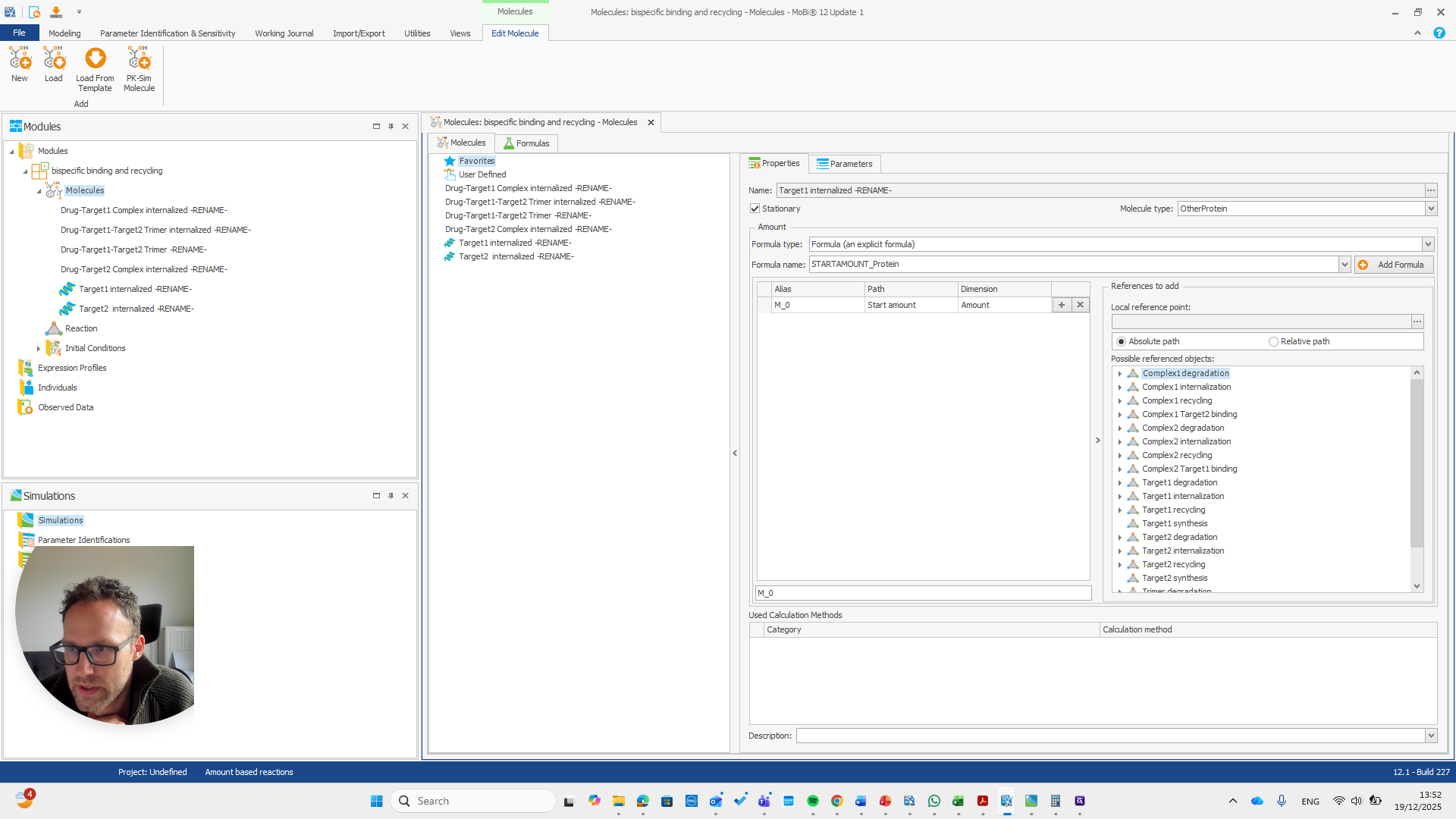
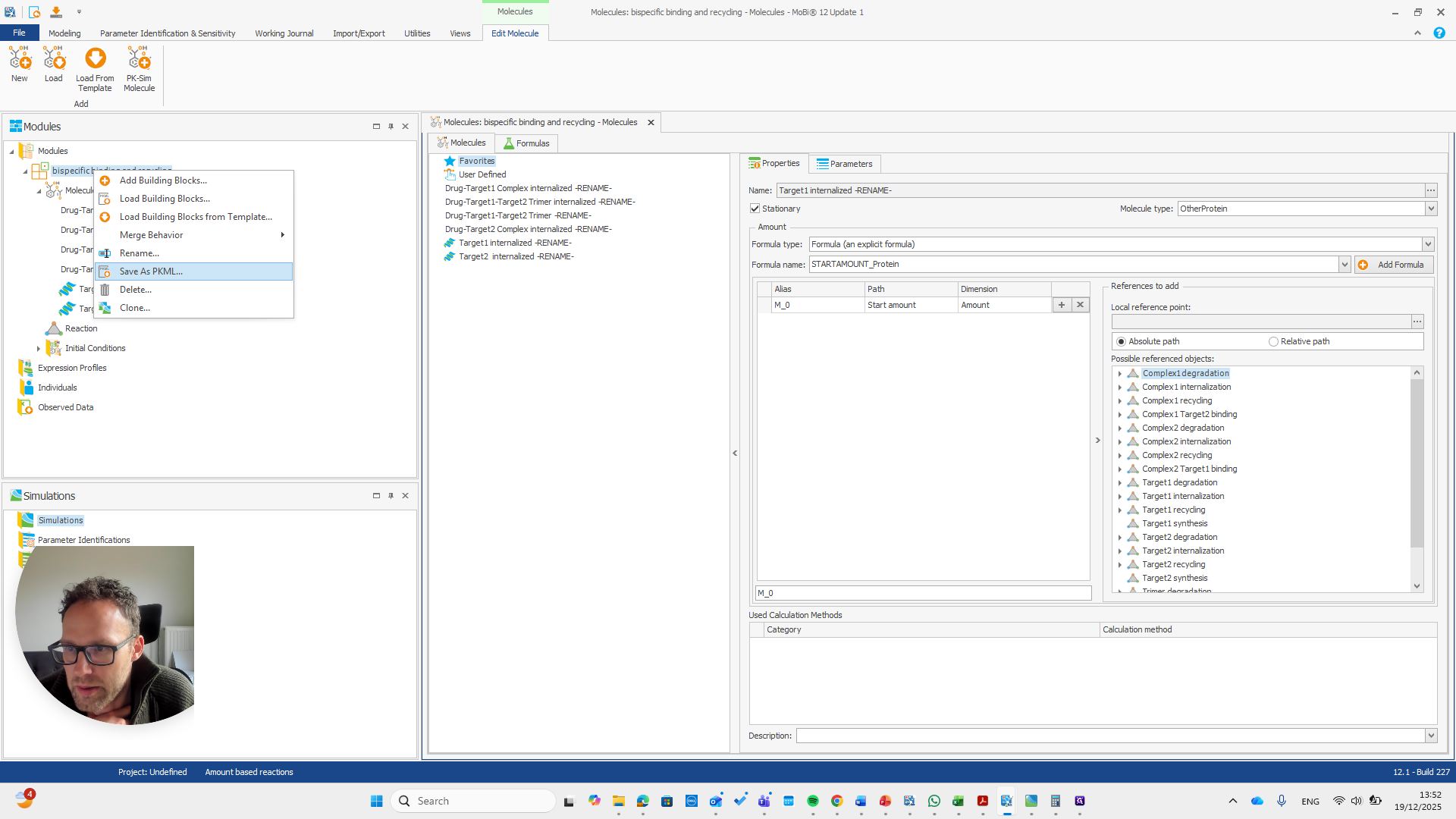
Now, we can return to our original file and load the PKML module.
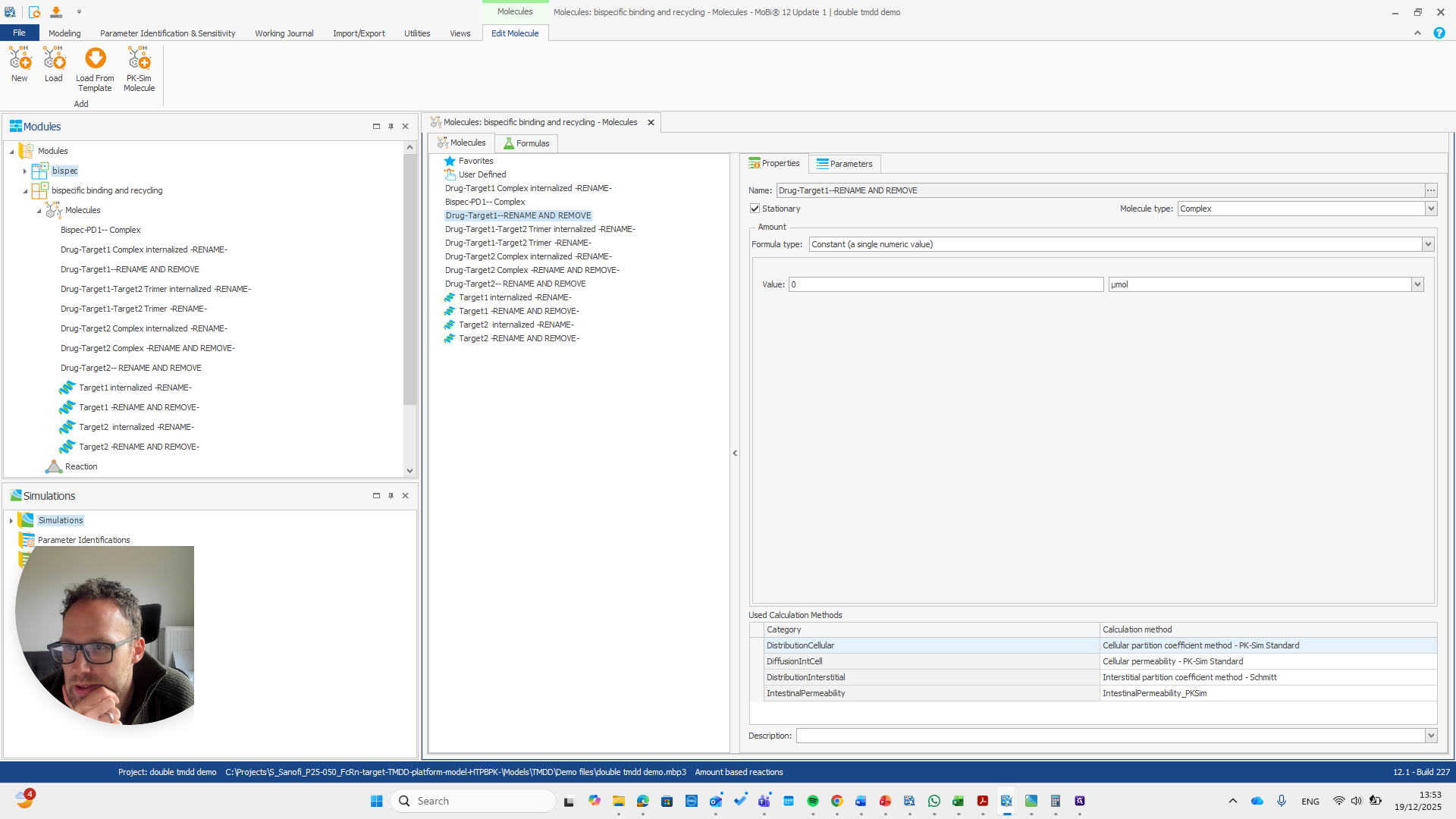
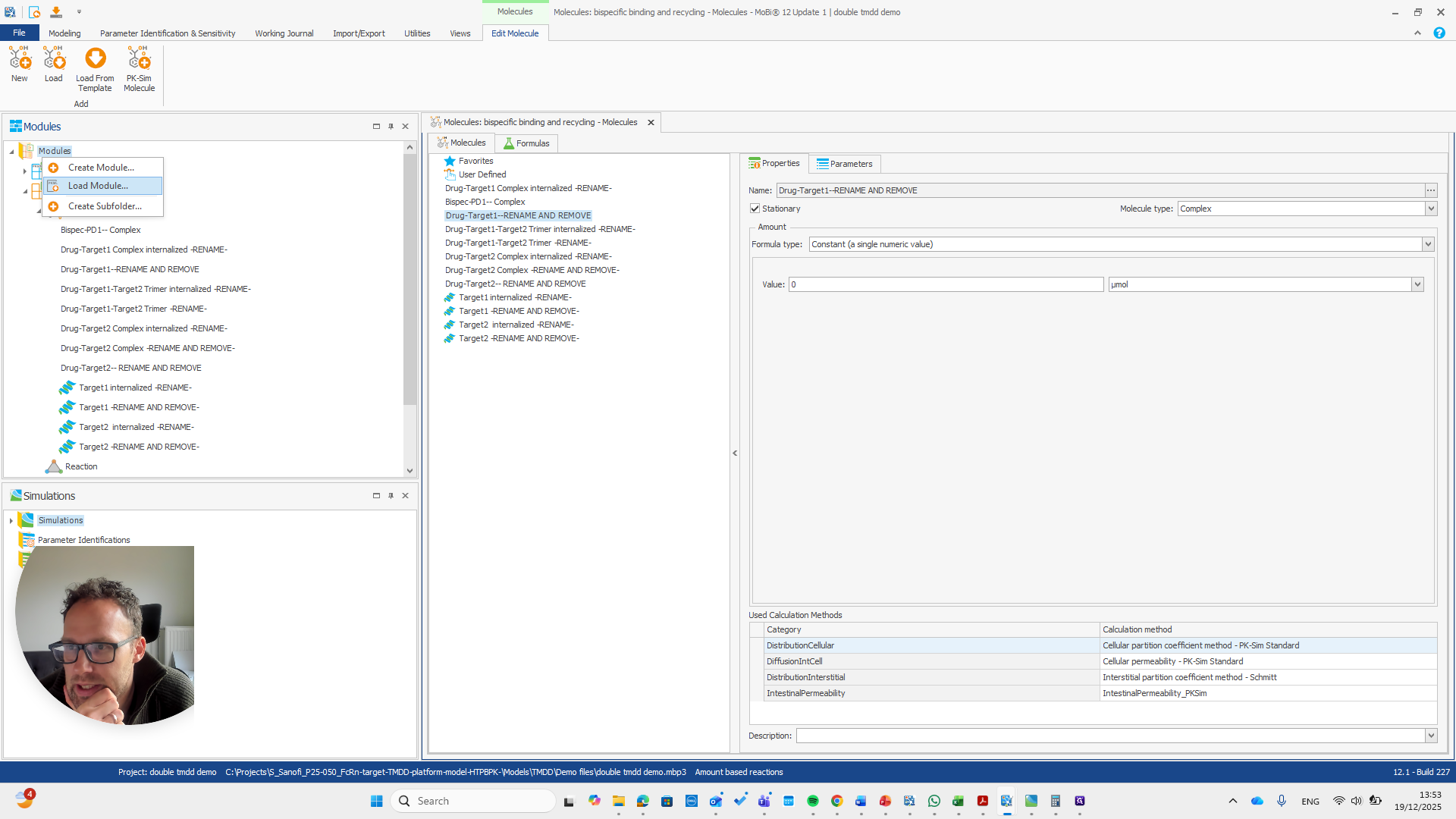
Alright. Now we can build a simulation using these modules.
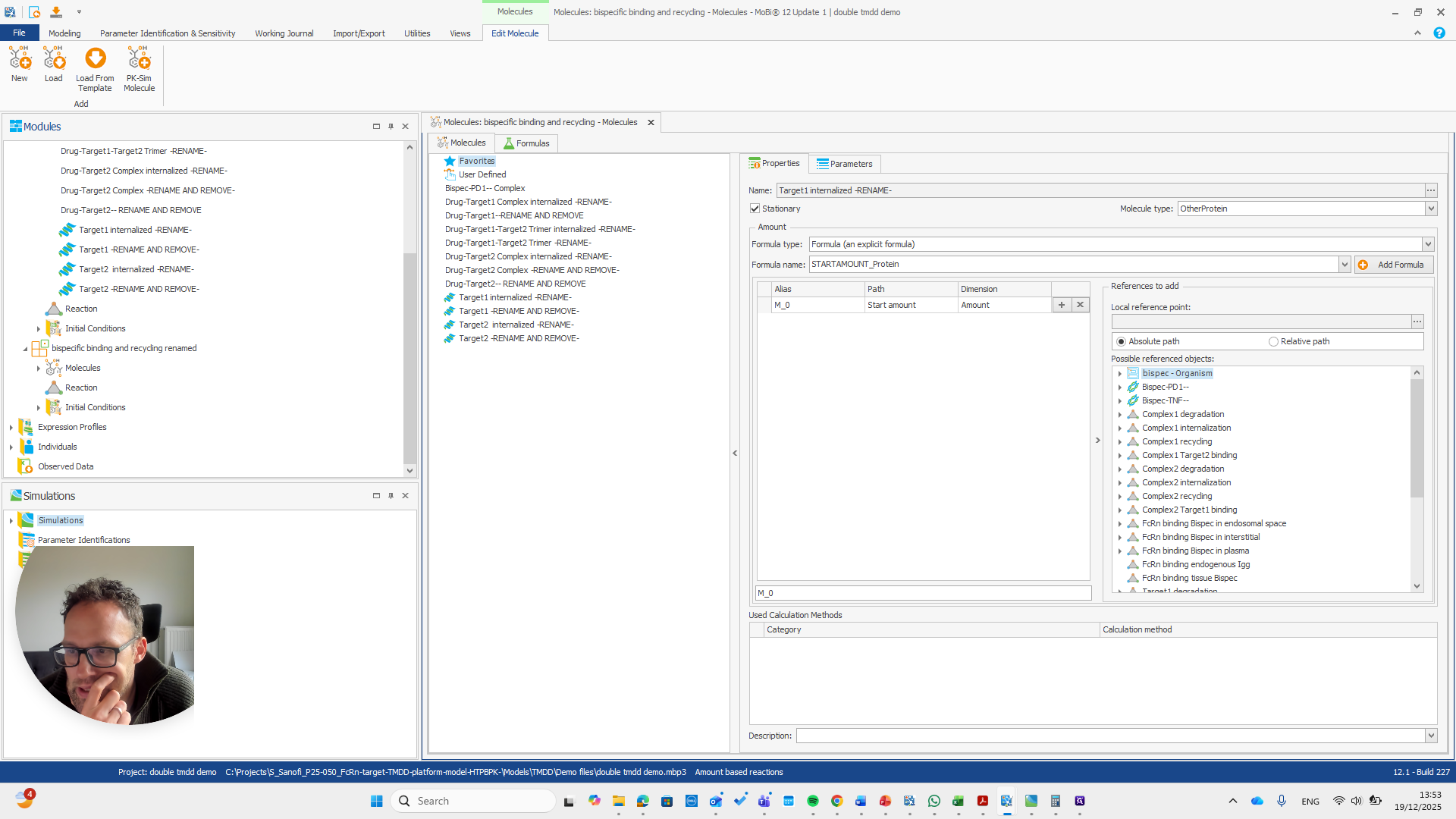
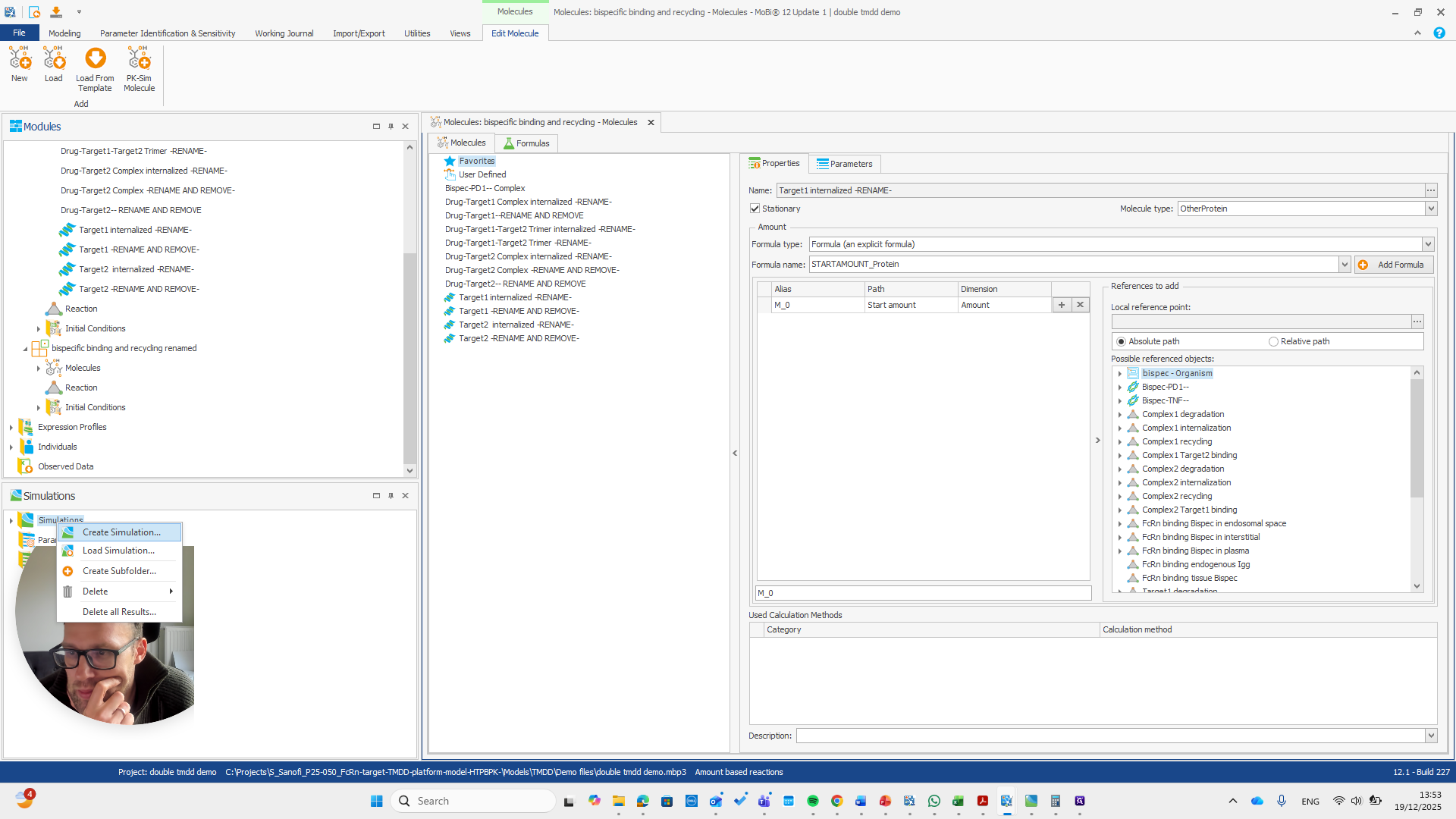
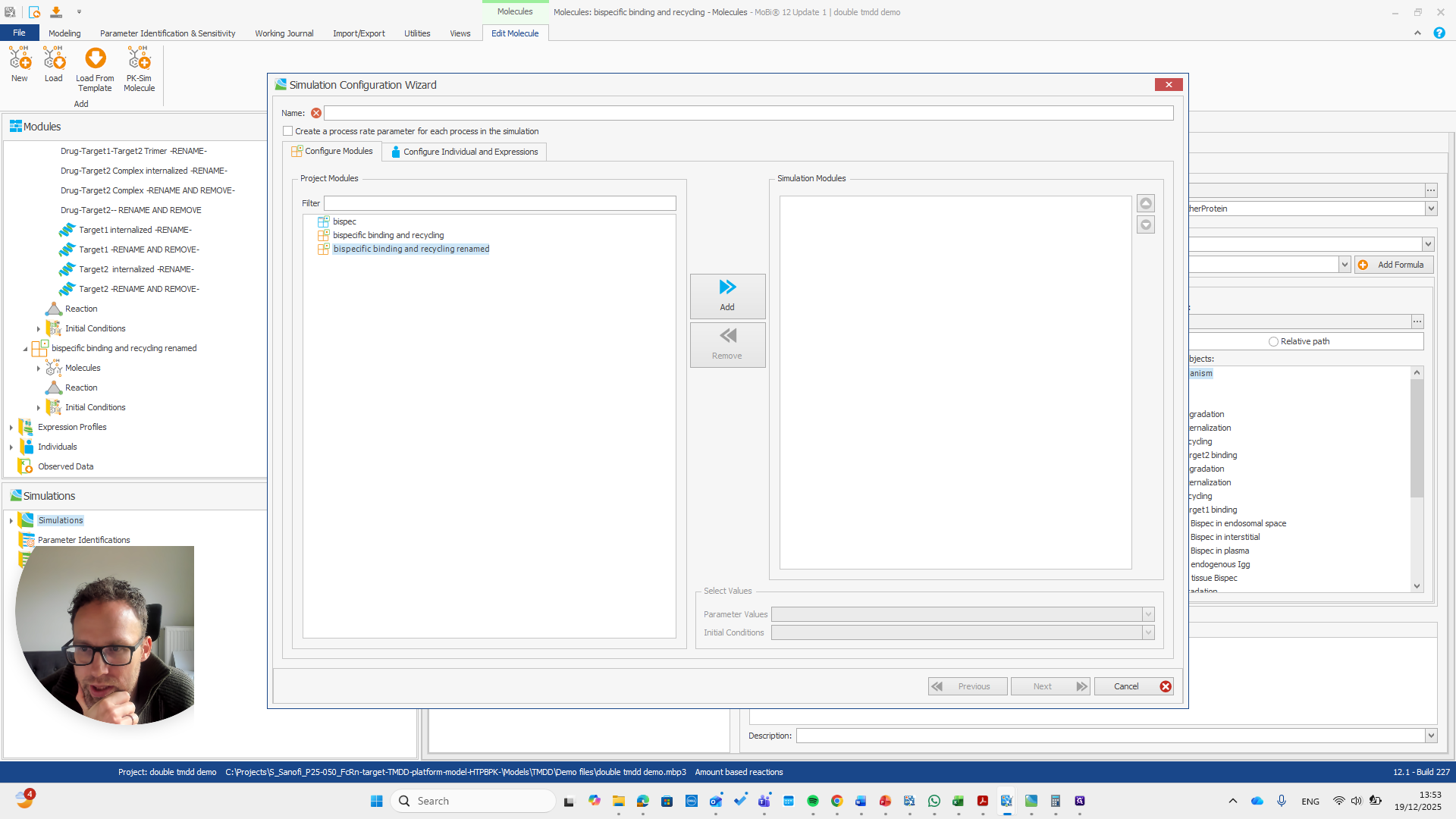
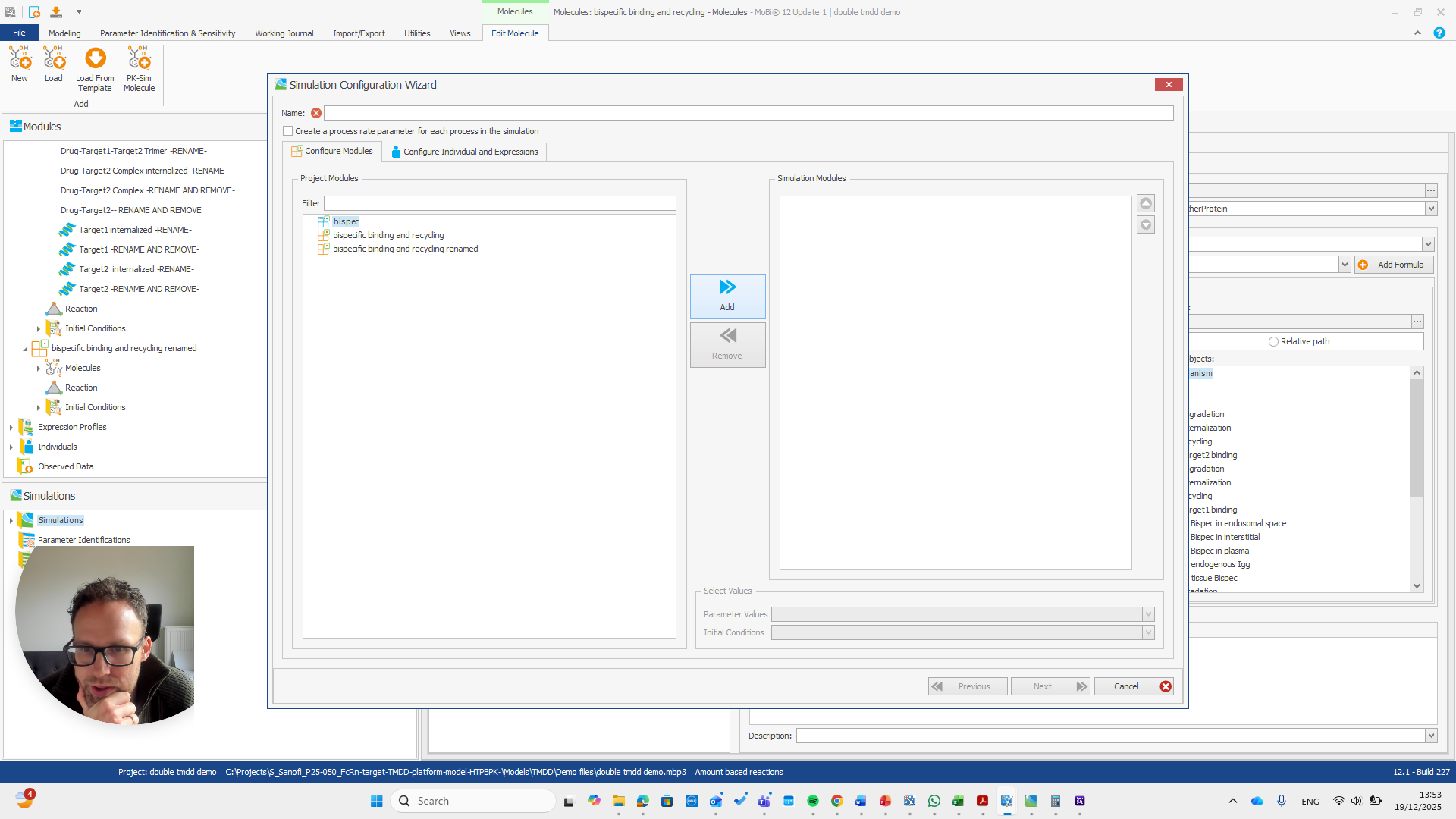
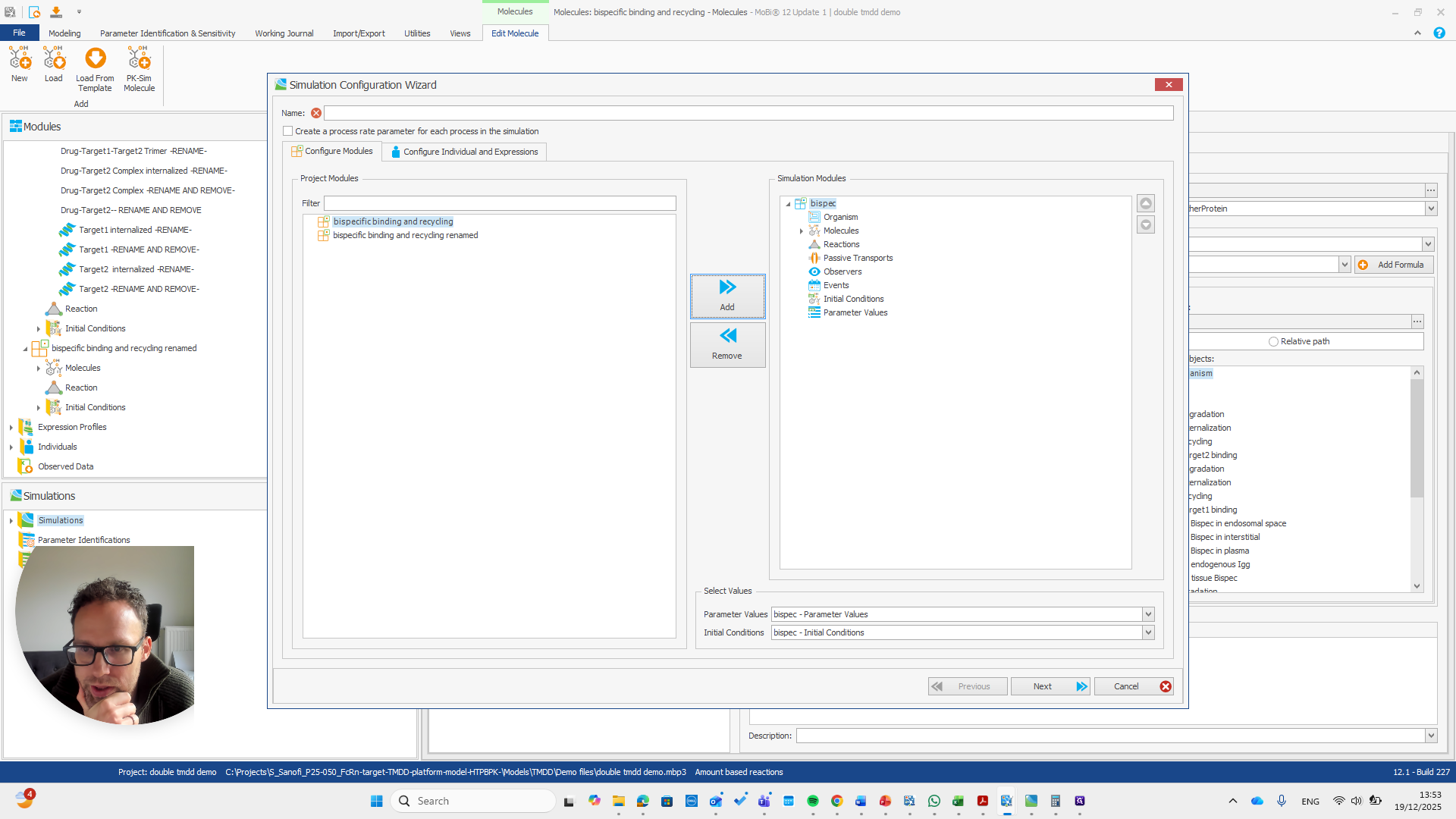
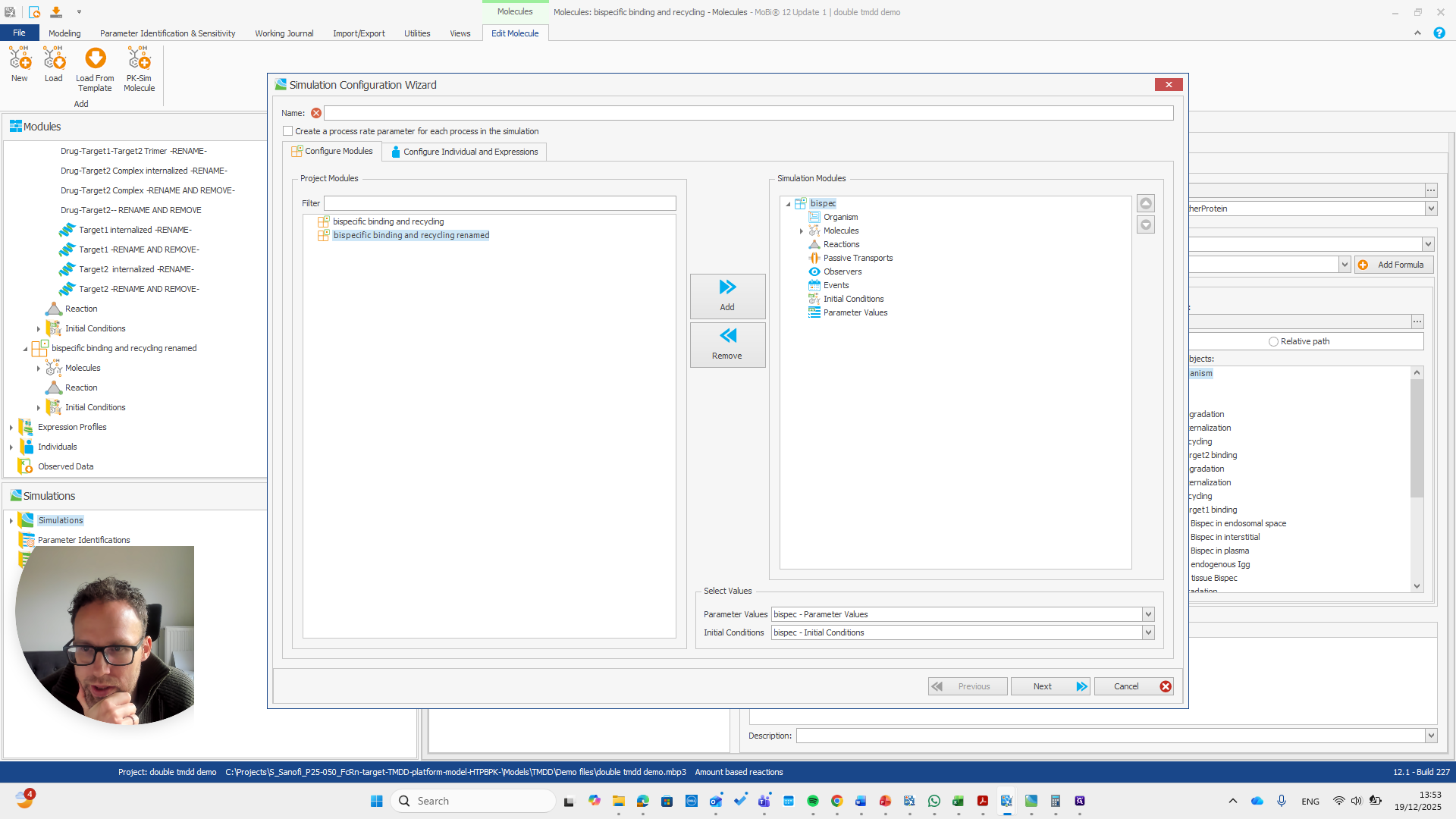
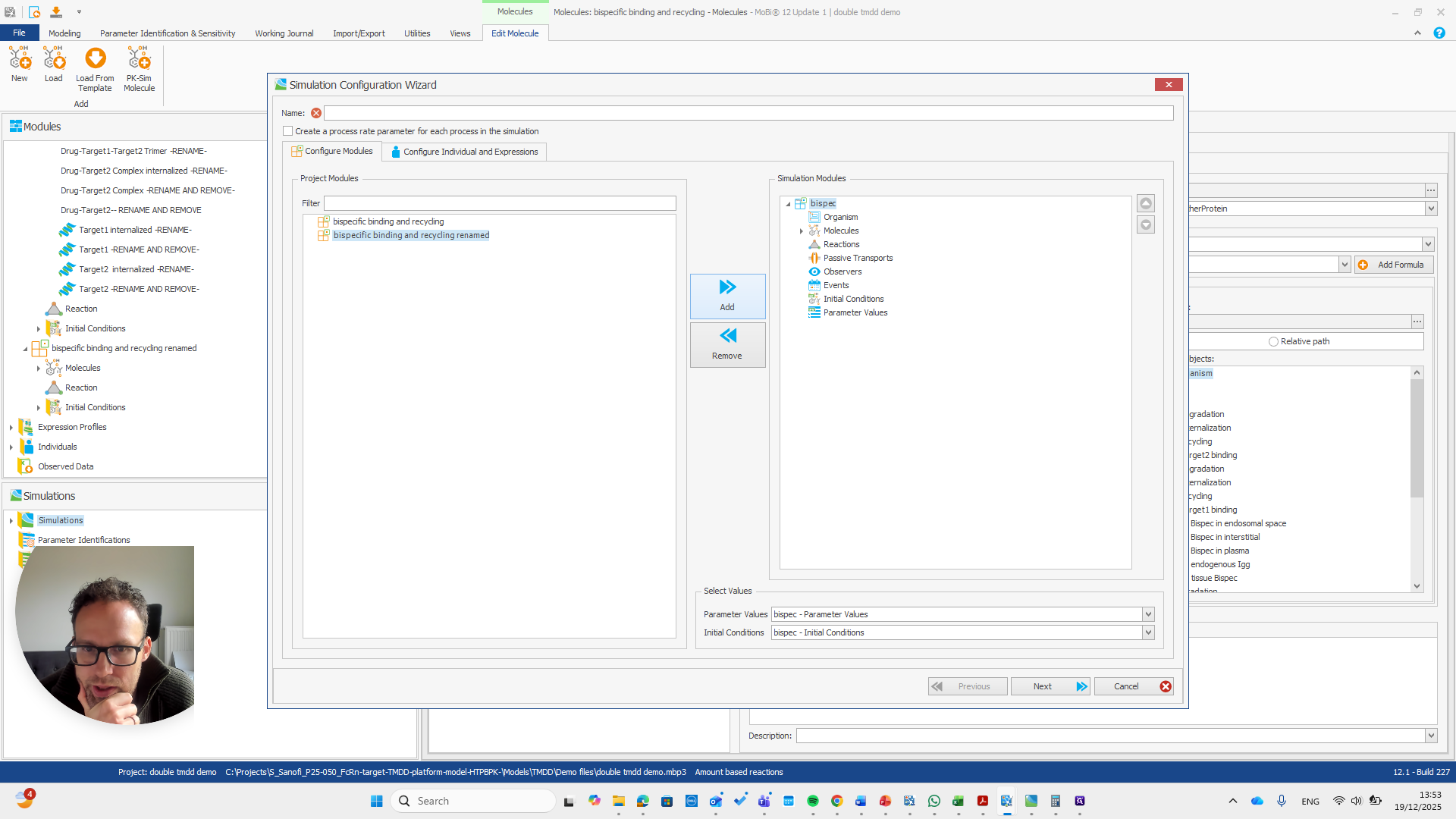
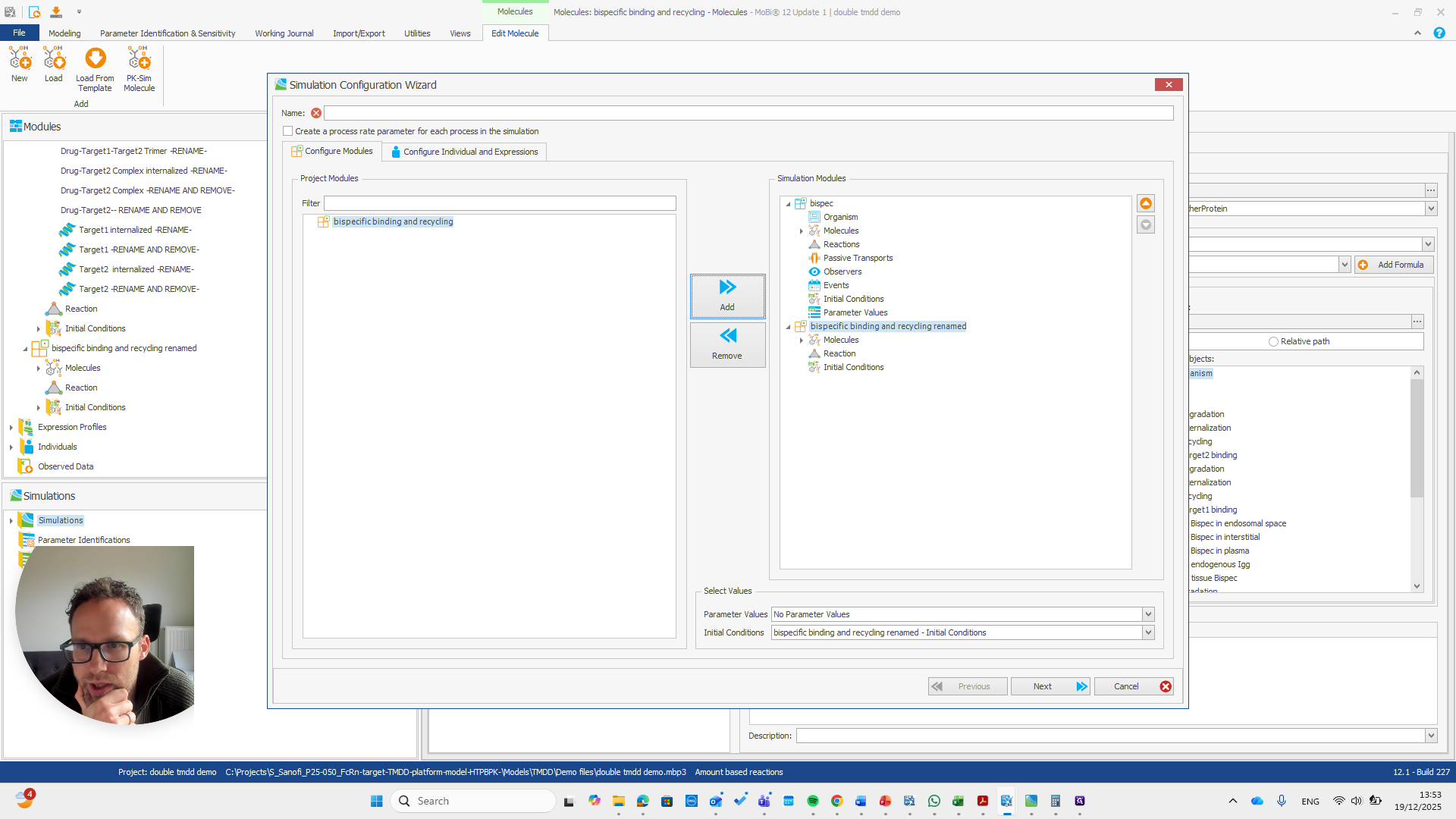
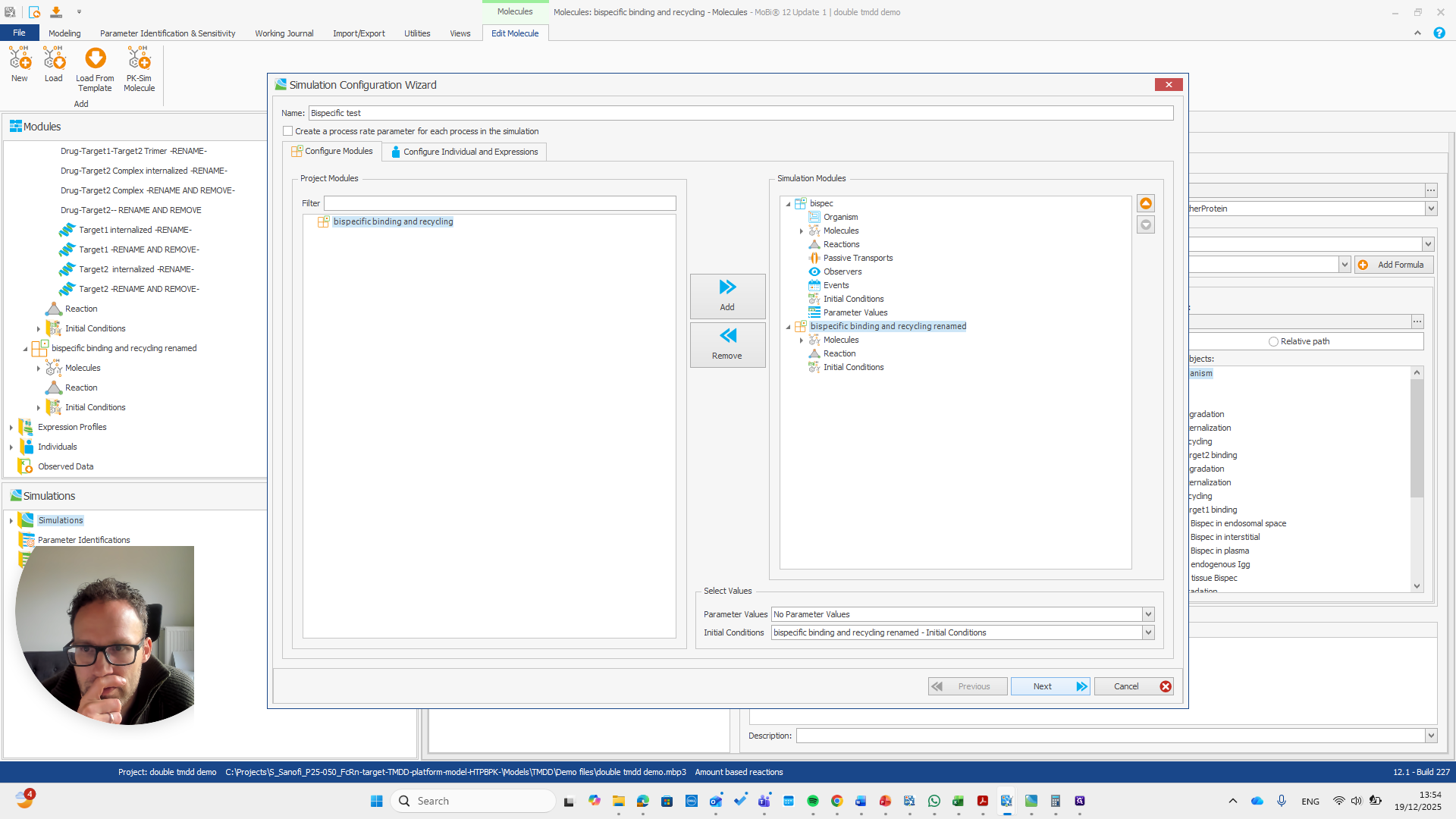
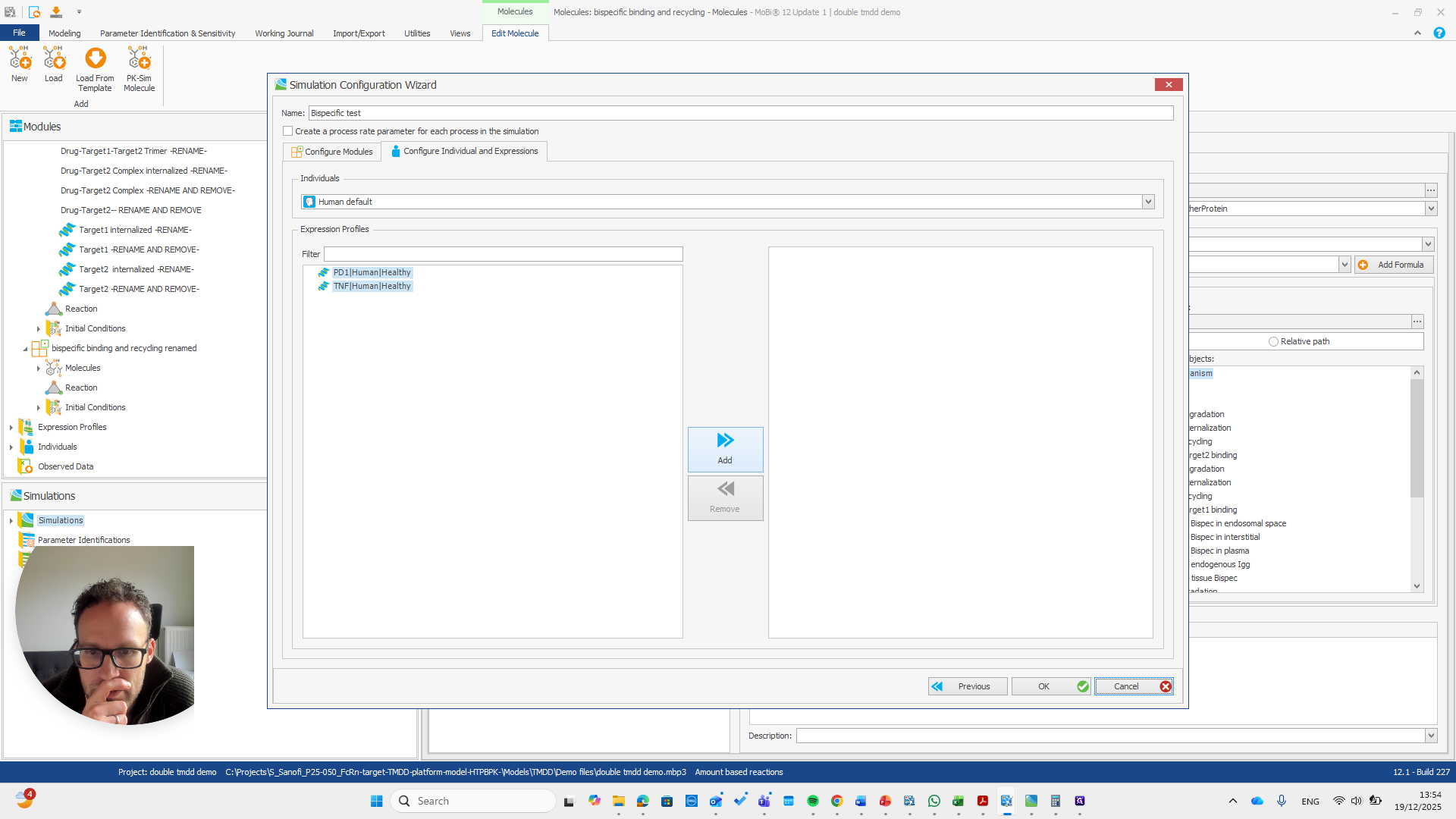
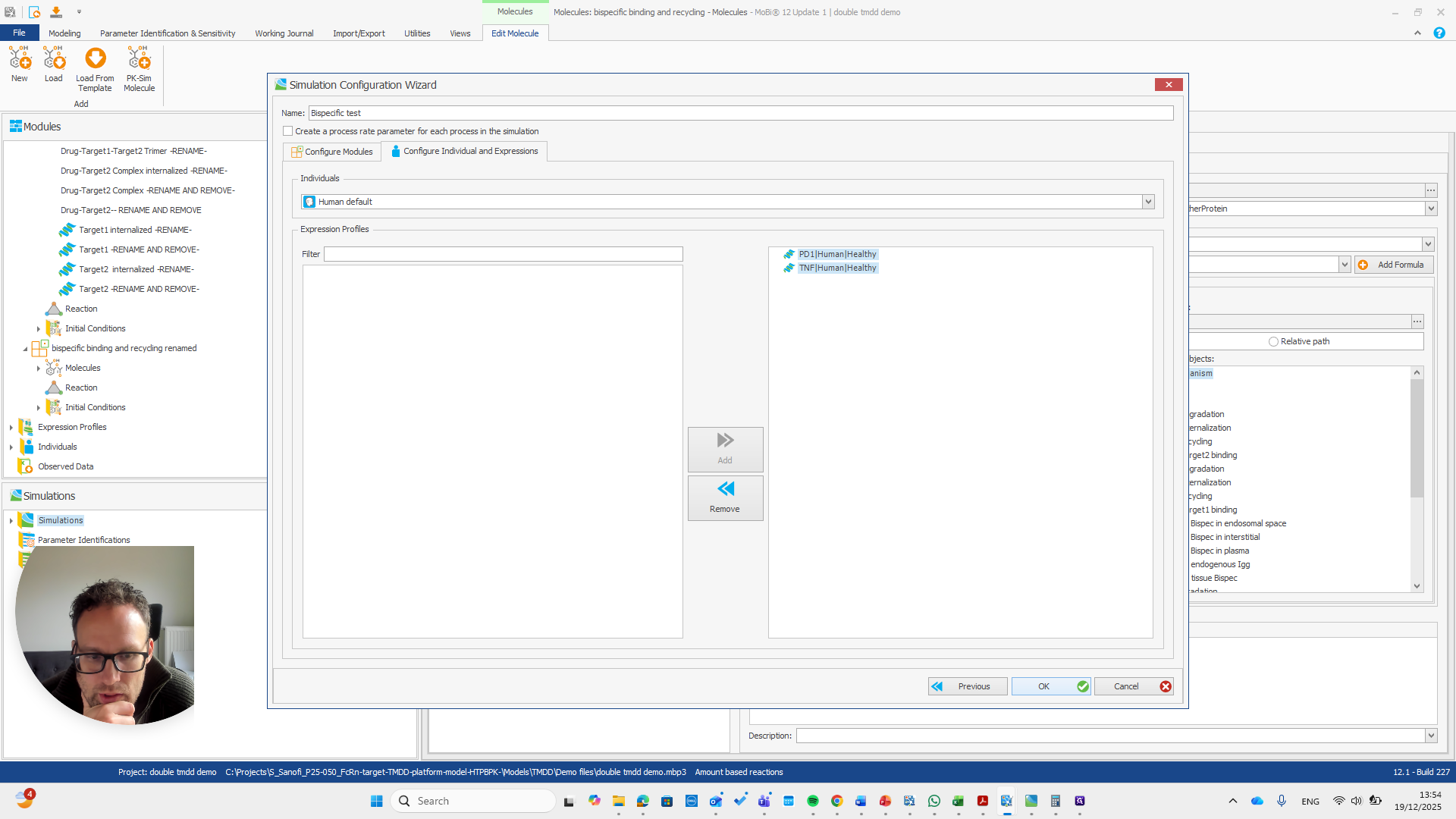
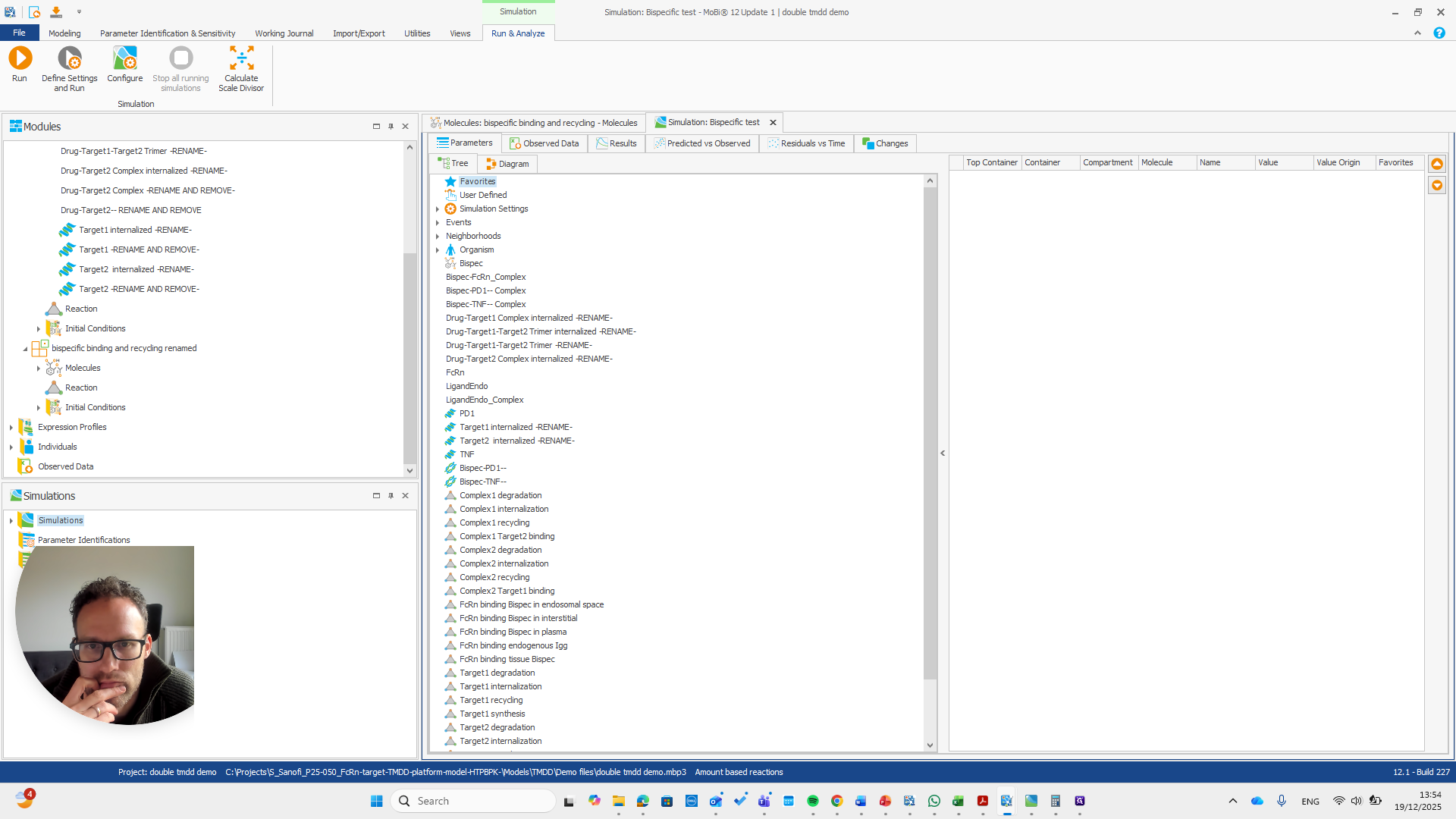
Okay. That seems to work. We do not receive any errors. We can run a simulation and perform all the quality checks we need.
We now have a working model, which means we also have a functioning module. Here, you can quickly check if all the reactions you added to the module appear in this list.